Volume 28, Number 6—June 2022
Dispatch
Introduction and Rapid Spread of SARS-CoV-2 Omicron Variant and Dynamics of BA.1 and BA.1.1 Sublineages, Finland, December 2021
Figure 2
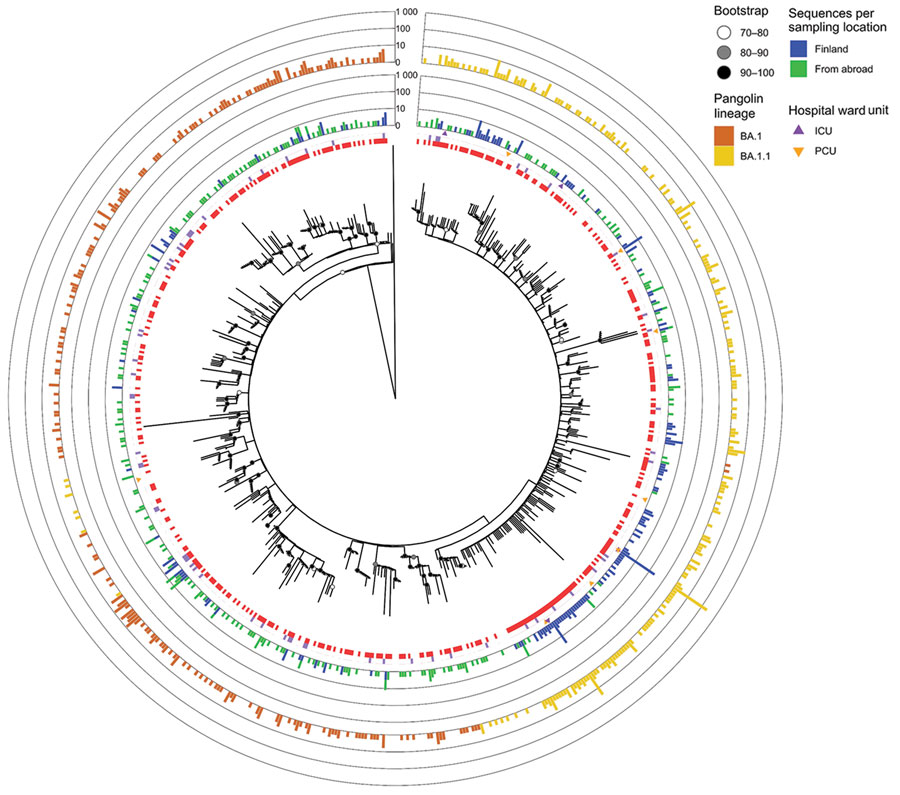
Figure 2. Clustering analysis of Omicron sequences in study of SARS-CoV-2 Omicron variant in Finland in late 2021–early 2022. The collapsed maximum-likelihood phylogenetic tree shows Omicron genomes sampled in Finland (n = 870) and reference sequences from other countries (n = 754), the reference dataset we used. The outermost bar plot shows the number of BA.1 and BA.1.1 sequences in each cluster. Purple squares indicate Omicron sequences collected from a Finland border; clusters with border samples each contain 1–9 sequences. Clustering analysis revealed that, by the beginning of January 2022, aside from 1 major BA.1.1 cluster (n = 236, 27.1% of all cases in Finland during the study period, November 29, 2021–January 6, 2022), most (n = 634, 72.8% of cases) Omicron cases in Finland were either singletons or small clusters (≤30 sequences). The tree was inferred using the IQTREE2 version 2.0.6 (http://www.iqtree.org) using ModelFinder and 1,000 bootstraps were computed with the integrated Ultrafast bootstrap algorithm and the clusters (red squares) with TreeCluster version 1.0.3 (https://github.com/niemasd/TreeCluster) using an arbitrary branch length of 0.001 and support value of 70. Triangles indicate sequences recorded from patients in the ICU or PCU. The tree is rooted to an Omicron BA.2 sequence (Genbank accession no. OV698431.1). ICU, intensive care unit; PCU, pulmonary care unit