Genetically Diverse Highly Pathogenic Avian Influenza A(H5N1/H5N8) Viruses among Wild Waterfowl and Domestic Poultry, Japan, 2021
Kosuke Okuya, Junki Mine, Kaori Tokorozaki, Isshu Kojima, Mana Esaki, Kohtaro Miyazawa, Ryota Tsunekuni, Saki Sakuma, Asuka Kumagai, Yoshihiro Takadate, Yuto Kikutani, Tsutomu Matsui, Yuko Uchida, and Makoto Ozawa

Author affiliations: Kagoshima University, Kagoshima, Japan (K. Okuya, I. Kojima, M. Esaki, M. Ozawa); National Agriculture and Food Research Organization, Tsukuba, Japan (J. Mine, K. Miyazawa, R. Tsunekuni, S. Sakuma, A. Kumagai, Y. Takadate, Y. Uchida); Kagoshima Crane Conservation Committee, Izumi, Japan (K. Tokorozaki, T. Matsui, M. Ozawa); Ministry of Agriculture, Forestry and Fisheries, Tokyo, Japan (Y. Kikutani)
Main Article
Figure 2
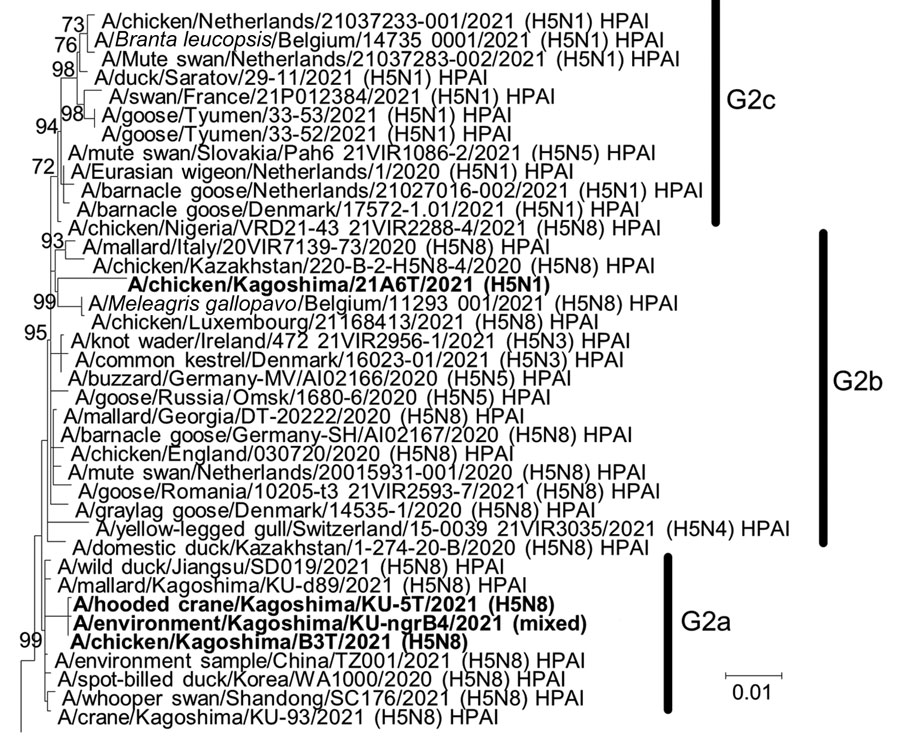
Figure 2. Phylogenetic tree of hemagglutinin genes of genetic group 2 (G2) of highly pathogenic avian influenza A(H5N1/H5N8) viruses isolated on the Izumi Plain, Japan, in November 2021. We phylogenetically analyzed the nucleotide sequences of the genes from A/environment/Kagoshima/KU-ngrB4/2021 (mixed), A/chicken/Kagoshima/21A6T/2021 (H5N1), A/chicken/Kagoshima/B3T/2021 (H5N8), and A/hooded crane/Kagoshima/KU-5T/2021 (H5N8) with representative counterparts by using the maximum-likelihood method with a bootstrapping set of 1,000 replicates. Bold text indicates viruses isolated in this study. Bootstrap values >70% are shown at the nodes. Scale bar indicates the number of nucleotide substitutions per site. HPAI, highly pathogenic avian influenza; LPAI, low pathogenicity avian influenza.
Main Article
Page created: May 10, 2022
Page updated: June 18, 2022
Page reviewed: June 18, 2022
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
