Volume 28, Number 7—July 2022
Research Letter
Circulation of Enterovirus D68 during Period of Increased Influenza-Like Illness, Maryland, USA, 2021
Abstract
We report enterovirus D68 circulation in Maryland, USA, during September–October 2021, which was associated with a spike in influenza-like illness. The characterized enterovirus D68 genomes clustered within the B3 subclade that circulated in 2018 in Europe and the United States.
In early July 2021, the United States began to relax COVID-19 infection control measures. As the number of COVID-19 cases began to fall, cases of influenza-like illness (Appendix Table 1) continued to be seen in the Johns Hopkins Hospital system (Baltimore, MD, USA) through October 2021 (Appendix Figure 1). Enterovirus/rhinovirus were detectable throughout the pandemic (1,2), but their positivity markedly increased to reach 20.7% (of all samples tested for enterovirus/rhinovirus) in October 2021, surpassing all other respiratory viruses (Appendix Figure 2) (2).
Enterovirus-D68 (EV-D68) was associated with a large outbreak of respiratory disease in children in North America in 2014 and was subsequently linked to the occurrence of acute flaccid myelitis (AFM) (3). After the 2014 outbreak, active surveillance of EV-D68 was implemented in many countries in Asia, Europe, Africa, and the Americas. Data obtained through surveillance during 2014–2018 suggested a biennial circulation cycle in Europe and North America (4,5). However, despite this expected biennial pattern, EV-D68 detection in 2020 was lower than anticipated, and limited cases were detected in the United States (6). This change in the circulation of EV-D68 in 2020 might have been secondary to the widespread mitigation measures for COVID-19. Of note, a recent study from 8 countries in Europe reported a rapid increase in EV-D68 infections during July 31–October 14, 2021, which coincided with a period of relaxed COVID-19 mitigation measures (7).
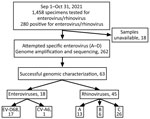
For this study, we collected samples positive for enterovirus/rhinovirus after the standard-of-care diagnosis at the Johns Hopkins Medical Microbiology Laboratory during September–October 2021 (Figure; Appendix). Research was conducted under Johns Hopkins Institutional Review Board protocol IRB00221396 with a waiver of consent. Remnant nasopharyngeal clinical specimens from patients that tested positive for enterovirus/rhinovirus during September–October 2021 were retrieved for the study. Genomes were made publicly available in GenBank (accession nos. OL826825–36).
We employed an optimized typing approach by using Nanopore next-generation sequencing (NGS) to characterize the enterovirus types (September–October 2021) associated with the increase in influenza-like illness. In brief, we used primers specific for enterovirus species A–D to amplify a ≈4,500-nt fragment that covers the whole P1 region (about half of the genome) (8) and then performed sequencing (Appendix). Of 280 enterovirus/rhinovirus-positive samples, we collected 262 for genotyping (Figure). We detected enterovirus in 28.6% of the 63 successfully sequenced samples (18/63); 94.4% (17/18) were EV-D68 and 5.6% (1/18) were coxsackievirus A6 (CV-A6). Even though the primers used for amplification were specific for enteroviruses, rhinoviruses were characterized in 45 of the 63 samples; those rhinoviruses consisted primarily of species C (26/45), followed by A (13/45) and B (6/45).
The whole cohort of patients positive for enterovirus/rhinovirus during September–October 2021 ranged in age from <1 year to >90 years; mean age was 16.7 years and median age 5 years. The male:female ratio was 1:1. On the other hand, the median age of EV-D68–positive patients was 2 years, and the male:female ratio was 1:3 (Appendix Table 2). EV-D68 was detected in 15/168 (8.9%) pediatric specimens positive for enterovirus/rhinovirus during the study time frame. Symptoms or signs of viral or respiratory illness were reported in all pediatric patients with EV-D68 (N = 15) (Appendix Table 2), and 5 patients (33.3%) were admitted and required supplemental oxygen, admission to the intensive care unit, or both. No neurologic complications including AFM were observed (Appendix Table 2). Of note, no AFM cases were diagnosed at Johns Hopkins Hospital during the study time frame. Most cases of enterovirus were detected in residents of the city of Baltimore (11/17). A total of 12 EV-D68 sequences, subclade B3, had a complete 5′ half of the genome (3000–4200 bp). EV-D68 genomes clustered with strains detected in 2019 from several countries in Europe (Appendix Figure 3).
We report a predominance of EV-D68 among the circulating enteroviruses during the same period in which enterovirus/rhinovirus positivity increased in this hospital system (2). The predominance of EV-D68 in our study (27% of total enterovirus/rhinovirus-typed genomes) was higher than the 0.4% and 3.6% observed in 2019 and 2020 in the United States (6) and comparable to the 24.3% reported before the COVID-19 pandemic in 2018 (6).
The EV-D68 strains detected belong to the B3 subclade, which had not been reported from the United States since 2018 (6) but was detected in Europe in 2019 (9). The strains we detected form a distinct cluster within the B3 subclade that circulated in 2018 in Europe and the United States but seem very close to those characterized in Europe in 2019. Nevertheless, it was reported that strains circulating in Europe in 2019 are common ancestors of strains detected in the United States in 2018 (9). That report might explain why the strains we identified are more closely related to subclade B3 from the United States than to those from Europe in 2018.
Dr. Fall is a postdoctoral research fellow in the Department of Pathology, Division of Medical Microbiology, Johns Hopkins School of Medicine. His primary research focus is respiratory viral surveillance, particularly enteroviruses and adenoviruses. Dr. Mostafa is an assistant professor of pathology and director of the Molecular Virology Laboratory at Johns Hopkins School of Medicine. Her research focuses on viral genomic evolution and its association with outbreaks and severe disease.
Acknowledgments
This publication was made possible by support from the Sherrilyn and Ken Fisher Center for Environmental Infectious Diseases, Division of Infectious Diseases, Johns Hopkins University School of Medicine. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of the Fisher Center or Johns Hopkins University School of Medicine.
H.H.M. is supported by the HIV Prevention Trials Network sponsored by the National Institute of Allergy and Infectious Diseases. Funding was provided by the Johns Hopkins Center of Excellence in Influenza Research and Surveillance (HHSN272201400007C), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research, of the NIH, DHHS (UM1 AI068613), the NIH RADx-Tech program (3U54HL143541-02S2), National Institute of Health RADx-UP initiative (grant R01 DA045556-04S1), Centers for Disease Control (contract 75D30121C11061), the Johns Hopkins University President’s Fund Research Response, the Johns Hopkins Department of Pathology, and the Maryland Department of Health. E.K. was supported by Centers for Disease Control and Prevention MInD-Healthcare Program (Grant no. U01CK000589).
References
- Olsen SJ, Winn AK, Budd AP, Prill MM, Steel J, Midgley CM, et al. Changes in influenza and other respiratory virus activity during the COVID-19 pandemic—United States, 2020–2021. MMWR Morb Mortal Wkly Rep. 2021;70:1013–9. DOIPubMedGoogle Scholar
- Uhteg K, Amadi A, Forman M, Mostafa HH. Circulation of Non-SARS-CoV-2 respiratory pathogens and coinfection with SARS-CoV-2 amid the COVID-19 pandemic. Open Forum Infect Dis. 2021;9:b618. DOIPubMedGoogle Scholar
- Aliabadi N, Messacar K, Pastula DM, Robinson CC, Leshem E, Sejvar JJ, et al. Enterovirus D68 infection in children with acute flaccid myelitis, Colorado, USA, 2014. Emerg Infect Dis. 2016;22:1387–94. DOIPubMedGoogle Scholar
- Kramer R, Sabatier M, Wirth T, Pichon M, Lina B, Schuffenecker I, et al. Molecular diversity and biennial circulation of enterovirus D68: a systematic screening study in Lyon, France, 2010 to 2016. Euro Surveill. 2018;23:
1700711 . DOIPubMedGoogle Scholar - Messacar K, Pretty K, Reno S, Dominguez SR. Continued biennial circulation of enterovirus D68 in Colorado. J Clin Virol. 2019;113:24–6. DOIPubMedGoogle Scholar
- Shah MM, Perez A, Lively JY, Avadhanula V, Boom JA, Chappell J, et al. Enterovirus D68-associated acute respiratory illness—new vaccine surveillance network, United States, July–November 2018–2020. MMWR Morb Mortal Wkly Rep. 2021;70:1623–8. DOIPubMedGoogle Scholar
- Benschop KS, Albert J, Anton A, Andrés C, Aranzamendi M, Armannsdóttir B, et al. Re-emergence of enterovirus D68 in Europe after easing the COVID-19 lockdown, September 2021. Euro Surveill. 2021;26:26. DOIPubMedGoogle Scholar
- Joffret ML, Polston PM, Razafindratsimandresy R, Bessaud M, Heraud JM, Delpeyroux F. Whole genome sequencing of enteroviruses species A to D by high-throughput sequencing: application for viral mixtures. Front Microbiol. 2018;9:2339. DOIPubMedGoogle Scholar
- Midgley SE, Benschop K, Dyrdak R, Mirand A, Bailly JL, Bierbaum S, et al. Co-circulation of multiple enterovirus D68 subclades, including a novel B3 cluster, across Europe in a season of expected low prevalence, 2019/20. Euro Surveill. 2020;25:25. DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: May 31, 2022
Table of Contents – Volume 28, Number 7—July 2022
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Heba Mostafa, The Johns Hopkins Hospital, Meyer B-121F, 600 N. Wolfe St, Baltimore, MD 21287, USA
Top