Volume 28, Number 9—September 2022
Research Letter
Tropheryma whipplei Intestinal Colonization in Migrant Children, Greece
Figure
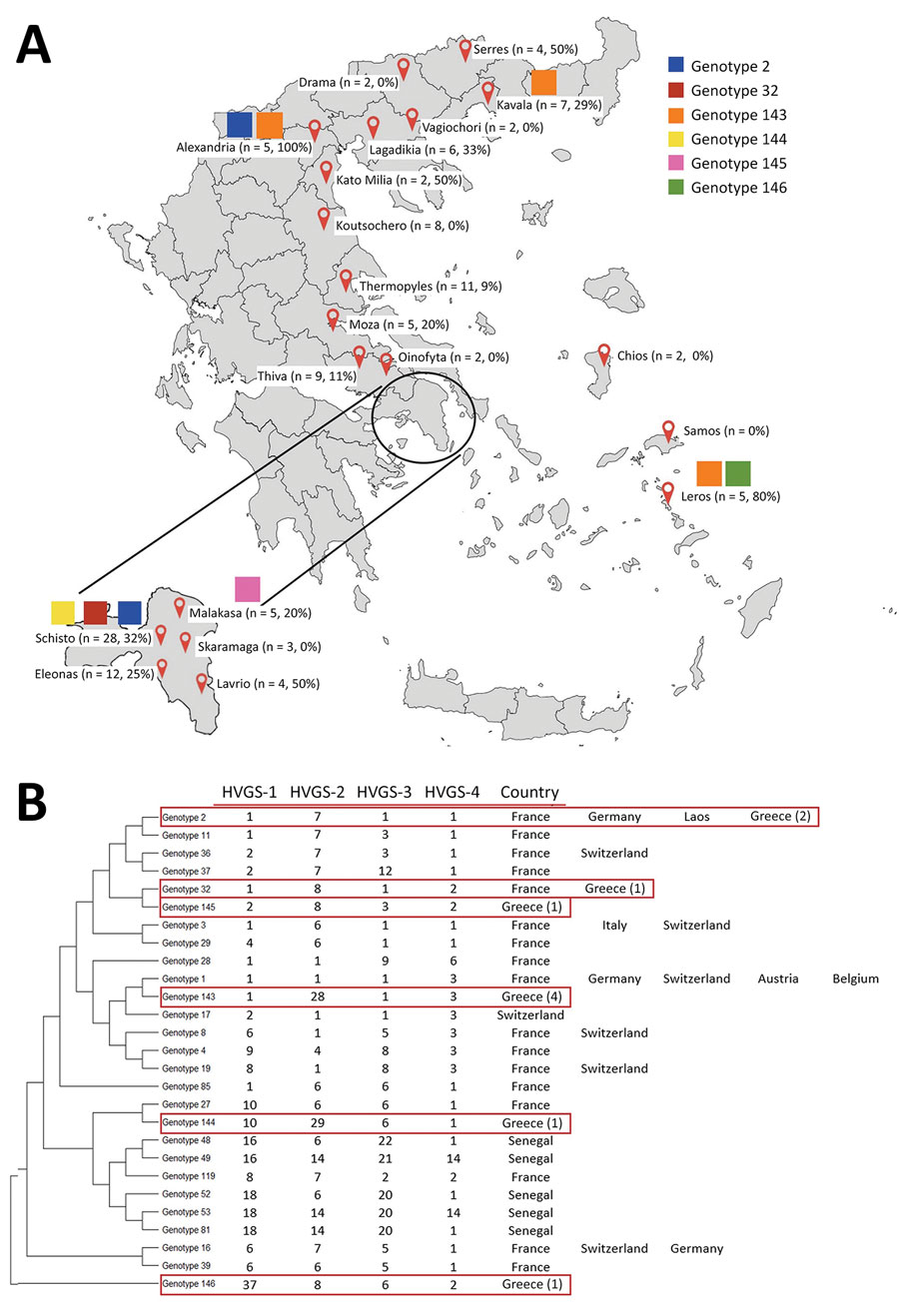
Figure. Results of stool sample tests for Tropheryma whipplei from migrant children 0–12 years of age from 20 hotspots throughout Greece. A) Defined hotspots throughout Greece, showing numbers and percentages of T. whipplei recovered from each location and distribution of different genotypes. B) Phylogenetic diversity of 6 genotypes of T. whipplei obtained from migrants (red boxes). Phylogenetic tree was constructed by using the maximum-likelihood method based on the Tamura 3-parameter substitution model. Sequences from the 4 HVGSs were concatenated. Noted next to the genotypes are the countries in which they have been previously detected. Numbers in parentheses note positive test results for children based on each genotype found in Greece. HVGS, highly variable genomic sequence.