Volume 28, Number 9—September 2022
Research Letter
Molecular Epidemiology of Blastomyces gilchristii Clusters, Minnesota, USA
Figure
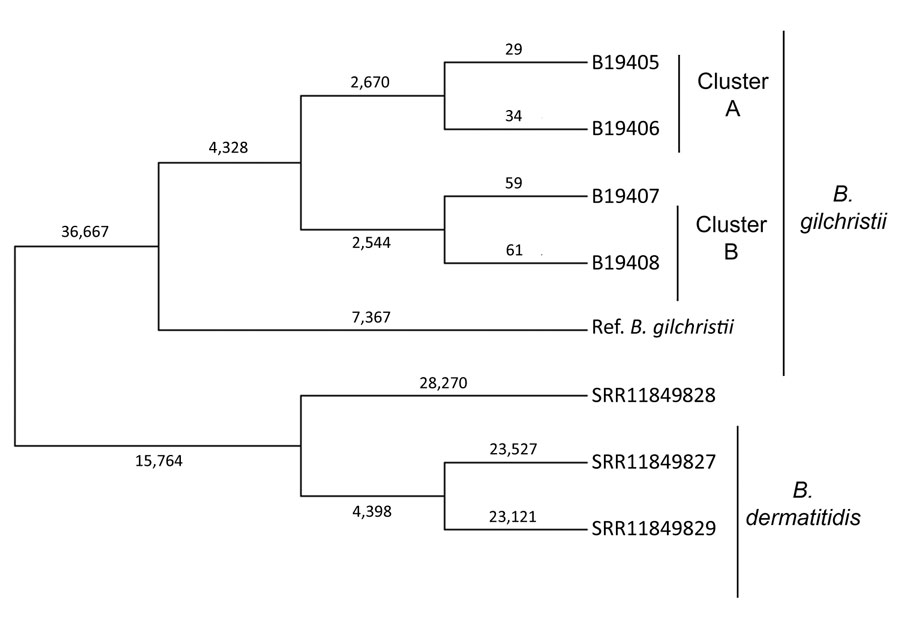
Figure. Genetic relationships and molecular epidemiology of Blastomyces gilchristii clusters, Minnesota, USA. We performed whole-genome sequencing of isolates from 4 patients in Minnesota who had Blastomyces gilchristii infections and compared the sequences with 3 publicly available B. dermatitidis isolates (National Center for Biotechnology Information run nos. SRR11849827, SRR11849828, SRR11849829). We analyzed single-nucleotide polymorphisms (SNPs) using the MycoSNP version 0.19 analytical workflow (https://github.com/CDCgov/mycosnp). We used the genome assembly data for B. gilchristii strain SLH14081 from GenBank (accession no. GCA_000003855.2) as a reference. Neighbor-joining tree shows the genetic relationships between cluster A and B, which each comprised isolates from 2 patients, the B. gilchristii reference strain, and B. dermatitidis isolates. Numbers represent the SNPs for each strain. Ref., reference.