Genome-Based Characterization of Listeria monocytogenes, Costa Rica
María Giralt-Zúñiga
12, Mauricio Redondo-Solano
1, Alexandra Moura
1, Nathalie Tessaud-Rita, Hélène Bracq-Dieye, Guillaume Vales, Pierre Thouvenot, Alexandre Leclercq, Carolina Chaves-Ulate, Kattia Núñez-Montero
3, Rossy Guillén-Watson, Olga Rivas-Solano, Grettel Chanto-Chacón, Francisco Duarte-Martínez, Vanessa Soto-Blanco, Javier Pizarro-Cerdá, and Marc Lecuit

Author affiliations: Institut Pasteur, Université Paris Cité, Inserm U1117, Biology of Infection Unit, National Reference Center, and WHO Collaborating Center Listeria, Paris, France (M. Giralt-Zúñiga, M. Redondo-Solano, A. Moura, N. Tessaud-Rita, H. Bracq-Dieye, G. Vales, P. Thouvenot, A. Leclercq, J. Pizarro-Cerdá, M. Lecuit); Instituto Tecnológico de Costa Rica, Cartago, Costa Rica (M. Giralt-Zúñiga, K. Núñez-Montero, R. Guillén-Watson, O. Rivas-Solano); University of Costa Rica, San José, Costa Rica (M. Redondo-Solano, C. Chaves-Ulate); Instituto Costarricense de Investigación y Enseñanza en Nutrición y Salud, Tres Rios, Costa Rica (G. Chanto-Chacón, F. Duarte-Martínez); National Animal Health Service, Heredia, Costa Rica (V. Soto-Blanco); Necker Enfants Malades University Hospital, APHP, Institut Imlagine, Paris (M. Lecuit)
Main Article
Figure
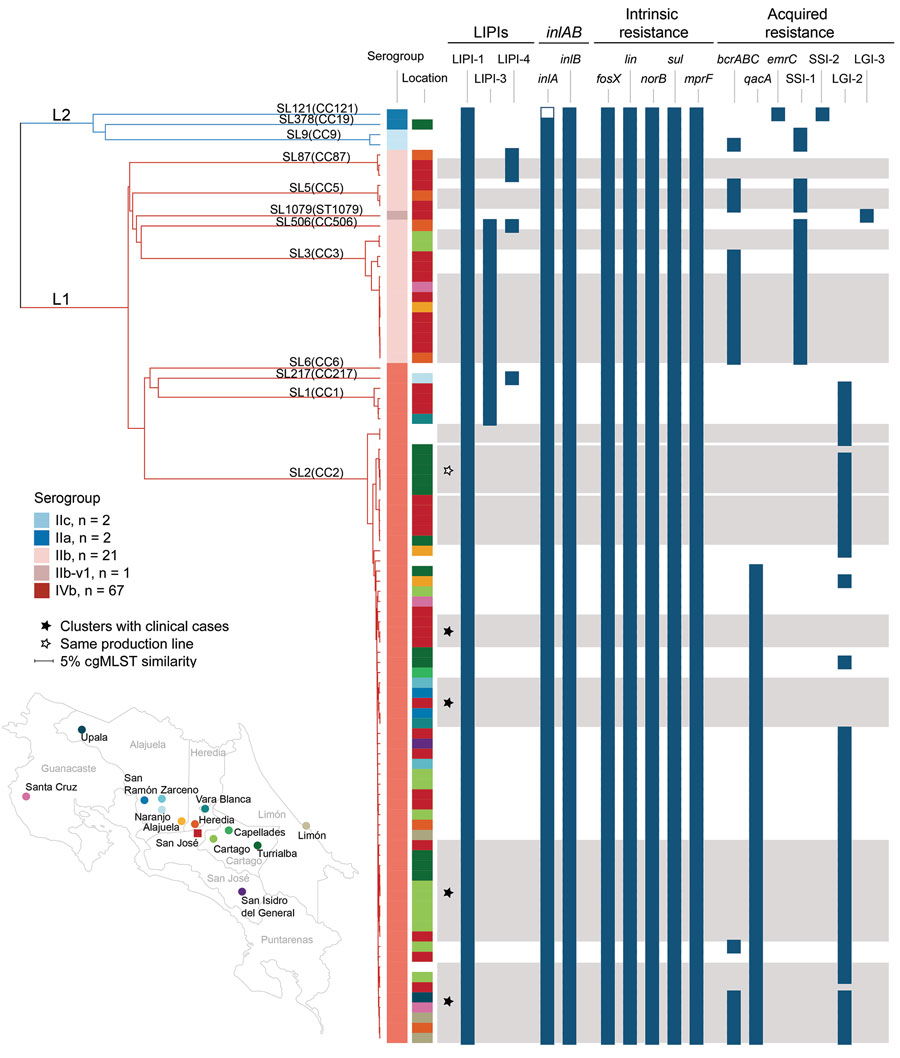
Figure. Single linkage dendrogram of 92 isolates generated for genome-based characterization of Listeria monocytogenes, Costa Rica. Dendrogram is based on core-genome multilocus sequence typing; (cgMLST) allelic profiles (1,748-locus scheme). Branches are colored according to lineages: L1, red; L2, blue. Branches are labeled according to lineages, sublineages, and clonal complexes. Information on isolates’ serogroup, and resistance profiles are provided in the columns. Colors in location column correspond to dots on map. Gray bars indicate clusters of isolates with <7 allelic differences out of 1,748 cgMLST loci. Presence of selected virulence and resistance genetic traits in each isolate is represented by dark blue boxes and empty boxes denote genes with premature stop codons. More details are provided in Appendix Figure 1. CC, clonal complex; L, lineage; LIPI, listeria pathogenicity island; SL, sublineage.
Main Article
Page created: October 13, 2023
Page updated: November 18, 2023
Page reviewed: November 18, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
