Molecular Detection and Characterization of Mycoplasma spp. in Marine Mammals, Brazil
Aricia Duarte-Benvenuto

, Carlos Sacristán, Ana Carolina Ewbank, Roberta Zamana-Ramblas, Henrique Christino Lial, Samira Costa Silva, Maria Alejandra Arias Lugo, Lara B. Keid, Caroline F. Pessi, José Rubens Sabbadini, Vanessa L. Ribeiro, Rodrigo del Rio do Valle, Carolina Pacheco Bertozzi, Adriana Castaldo Colosio, Hernani da Cunha Gomes Ramos, Angélica María Sánchez-Sarmiento, Raquel Beneton Ferioli, Larissa Pavanelli, Joana Midori Penalva Ikeda, Vitor L. Carvalho, Felipe Alexandre Catardo Gonçalves, Pablo Ibáñez-Porras, Irene Sacristán, and José Luiz Catão-Dias
Author affiliations: University of São Paulo, São Paulo, Brazil (A. Duarte-Benvenuto, A.C. Ewbank, R. Zamana-Ramblas, H.C. Lial, S.C. Silva, M.A.A. Lugo, L.B. Keid, J.L. Catão-Dias); Centro de Investigación en Sanidad Animal, Madrid, Spain (C. Sacristán, P. Ibáñez-Porras, I. Sacristán); Instituto de Pesquisas Cananéia, Cananéia, Brazil (C.F. Pessi, J.R. Sabbadini); Instituto Biopesca, Praia Grande, Brazil (V.L. Ribeiro, C.P. Bertozzi); Aiuká Consultoria Ambiental, Praia Grande (R. del Rio do Valle); Universidade Estadual Paulista, Campus do Litoral Paulista, São Vicente, Brazil (C.P. Bertozzi); Instituto Baleia Jubarte, Caravelas, Brazil (A. Castaldo Colosio, H. da Cunha Gomes Ramos); Instituto Argonauta para a Conservação Costeira e Marinha, Ubatuba, Brazil (A.M. Sánchez-Sarmiento, R. Beneton Ferioli); Instituto Mamíferos Aquáticos, Salvador, Brazil (L. Pavanelli, J.M. Penalva Ikeda); Associação de Pesquisa e Preservação de Ecossistemas Aquáticos, Caucaia, Brazil (V.L. Carvalho, F.A. Catardo Gonçalves)
Main Article
Figure 2
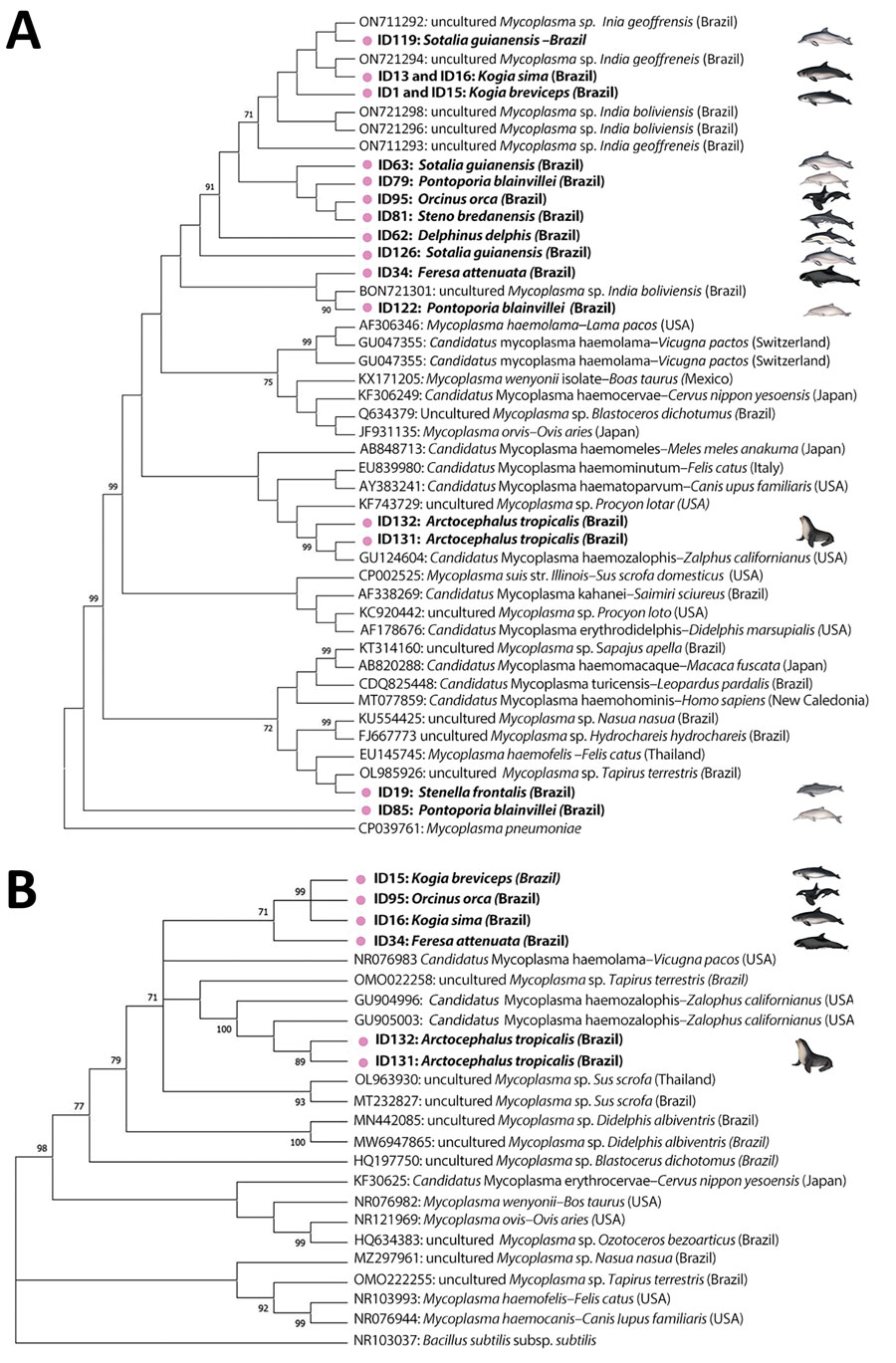
Figure 2. Maximum-likelihood phylograms from a study of molecular detection and characterization of Mycoplasma spp. in marine mammals, Brazil. A) 16S rRNA gene (329-bp) phylogram based on Tamura 3-parameter with inversions and gamma distribution model. Mycoplasma pneumoniae was selected as outgroup. B) 23S rRNA gene (820-bp) phylogram based on a general time-reversible model with invariant sites. Bacillus subtilis was selected as outgroup. Trees show alignment of Mycoplasma spp. consensus sequences obtained in marine mammals from this study (pink dots and bold text) and other hemotropic mycoplasmas retrieved from GenBank. Reliability of the phylograms was tested by 1,000 replicate bootstrap analyses omitting values <70. Trees generated by using MEGA 7.0 (https://www.megasoftware.net).
Main Article
Page created: October 13, 2023
Page updated: November 18, 2023
Page reviewed: November 18, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
