Volume 29, Number 2—February 2023
Research Letter
Metagenomic Sequencing of Monkeypox Virus, Northern Mexico
Figure
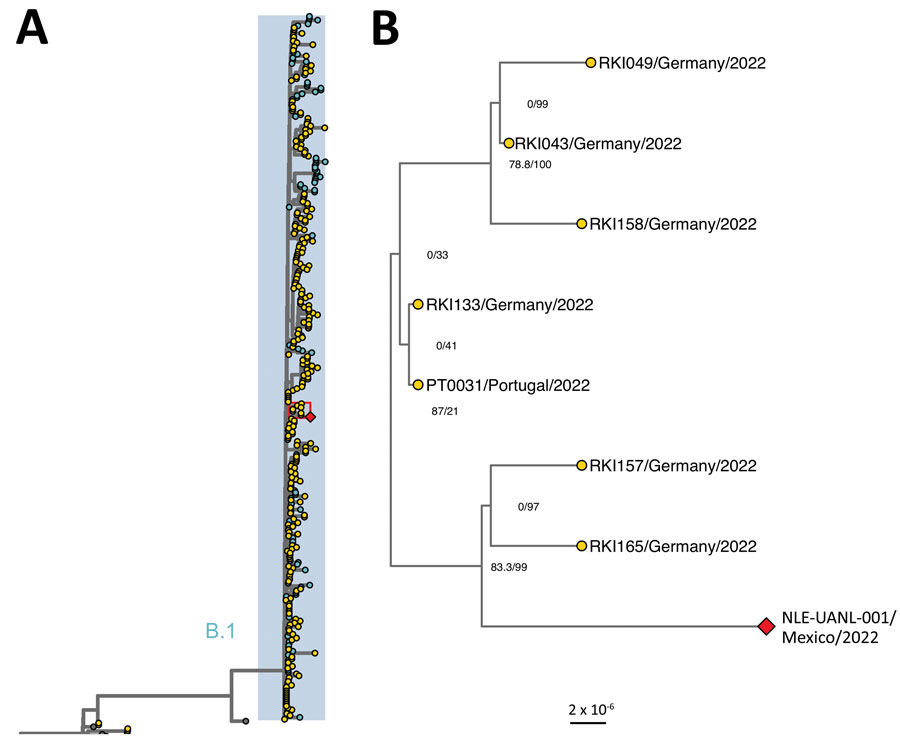
Figure. Phylogenetic relationship of monkeypox virus genomes, 2017–2022. A) Maximum-likelihood phylogenetic tree with 400 whole-genome sequences of monkeypox virus made in IQTree (http://www.iqtree.org). Highlighted genomes in blue belong to the proposed B.1 lineage. B) Magnification of area in red box in panel A showing clade indicating the Mexico isolate and related sequences. Node numbers indicate SH-like approximate likelihood ratio test and ultrafast bootstrap values. The red diamond corresponds to the sequence from this study and yellow circles to sequences from Europe. The list of accession numbers of the sequences is shown in the Appendix Table. The tree was annotated in ggtree (https://bioconductor.org/packages/release/bioc/html/ggtree.html). Scale bar indicates substitutions per site.