Volume 29, Number 3—March 2023
Research Letter
SARS-CoV-2 Infection in a Hippopotamus, Hanoi, Vietnam
Figure
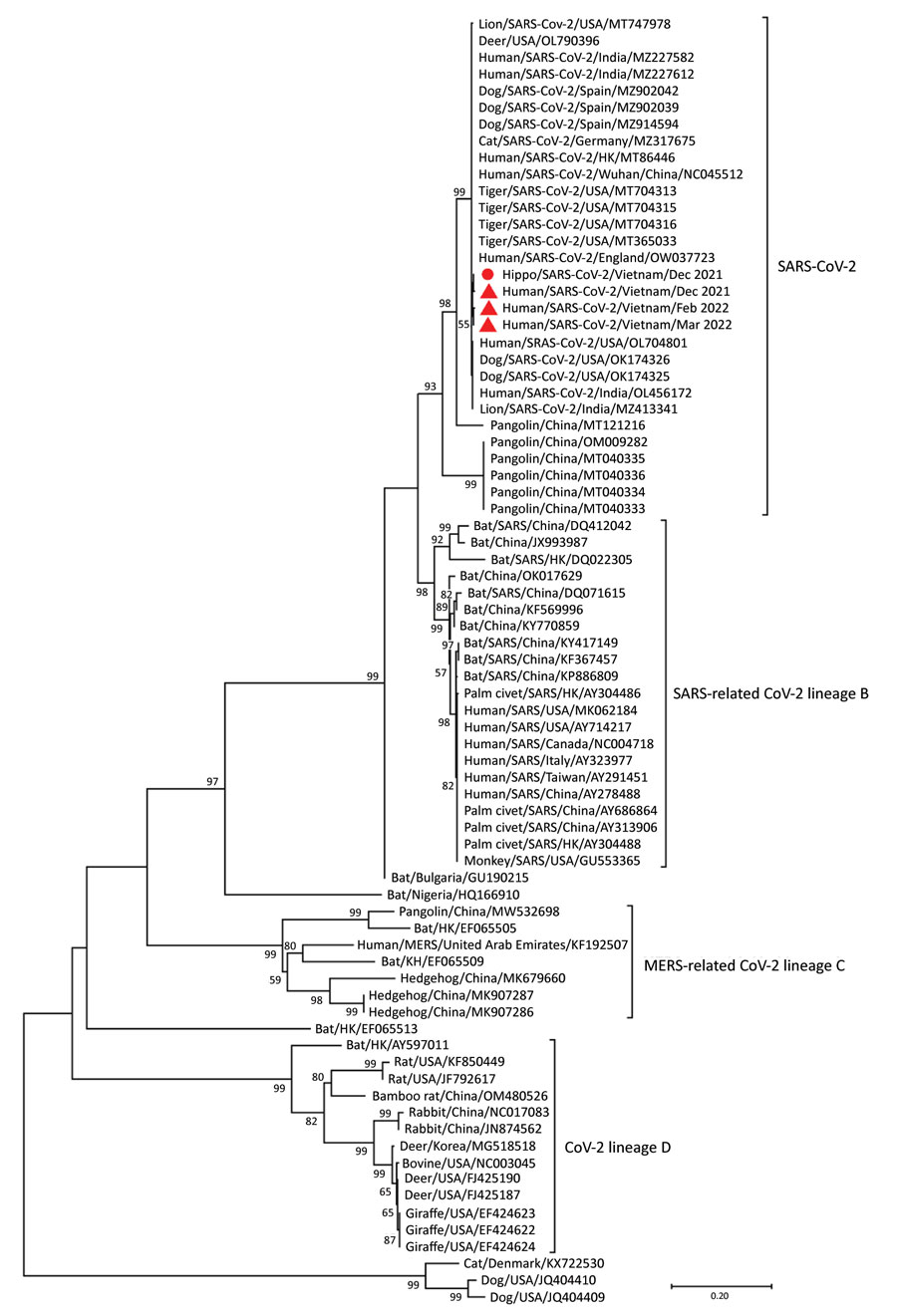
Figure. Maximum-likelihood tree constructed for 600-bp RNA-dependent RNA polymerase gene SARS-CoV-2 nucleotide sequences from a hippopotamus (red circle) and 3 human SARS-CoV-2 strains from COVID-19 patients in Vietnam (red triangles) compared with reference betacoronavirus strains obtained from GenBank. Betacoronavirus lineages are indicated on the right of the figure. Scale bar denotes evolutionary distance. MERS, Middle Eastern respiratory syndrome.
1These first authors contributed equally to this article.
Page created: January 12, 2023
Page updated: February 28, 2023
Page reviewed: February 28, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.