Volume 29, Number 3—March 2023
Dispatch
Reemergence of Lymphocytic Choriomeningitis Mammarenavirus, Germany
Figure
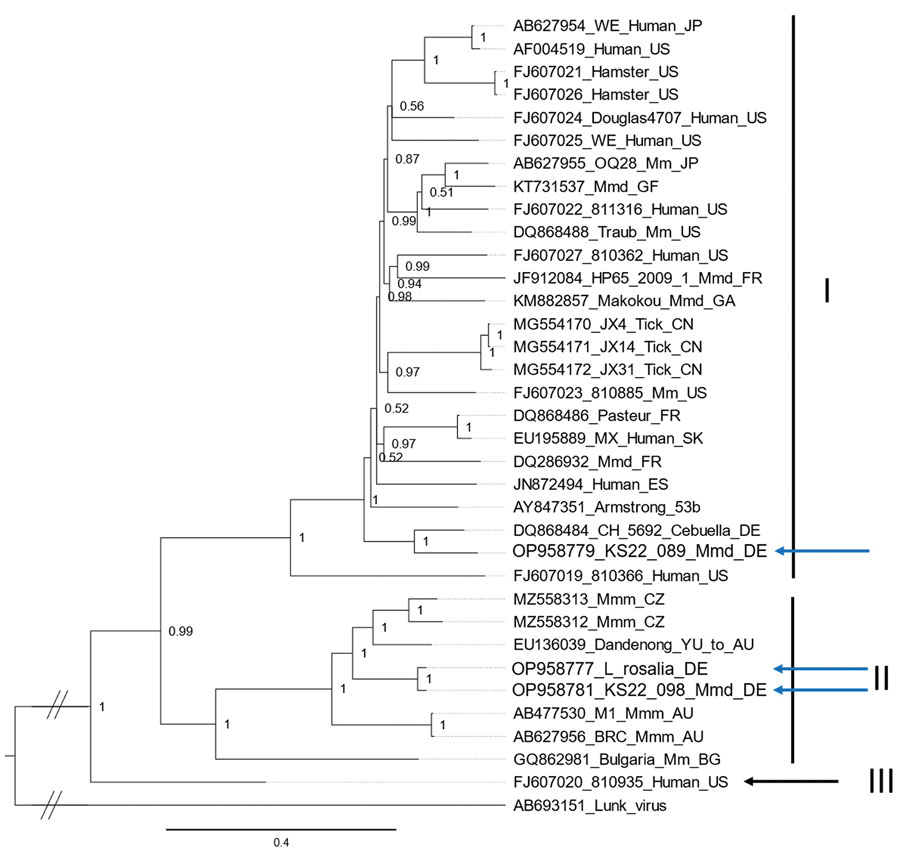
Figure. Phylogeny of the L protein encoding nucleotide sequences of lymphocytic choriomeningitis virus (LCMV) identified in Germany (blue arrows) and reference sequences, constructed by using Bayesian inference. Lunk virus from Mus minutoides mice was used as an outgroup. Sequence names are comprised of the GenBank accession number, strain name, host species and country of origin (wherever known). Countries are represented by their International Organization for Standardization code (AU, Australia; BG, Bulgaria; CN, China; DE, Germany; ES, Spain; FR, France; GA, Gabon; GF, French Guiana; JP, Japan; SK, Slovakia; US, USA; YU, former Yugoslavia). Roman numerals (I–IV) represent the different LCMV lineages defined according to Albariño et al. (2); WE and Armstrong refer to laboratory strains of LCMV. Mm, Mus musculus; Mmm, Mus musculus musculus; Mmd, Mus musculus domesticus.
References
- Meyer BJ. Arenaviruses: Genomic RNAs, transcription, and replication. In: Oldstone MBA, editor. Arenaviruses I. Berlin: Springer; 2002. p. 139–57.
- Albariño CG, Palacios G, Khristova ML, Erickson BR, Carroll SA, Comer JA, et al. High diversity and ancient common ancestry of lymphocytic choriomeningitis virus. Emerg Infect Dis. 2010;16:1093–100. DOIPubMedGoogle Scholar
- Armstrong C, Lillie RD. Experimental lymphocytic choriomeningitis of monkeys and mice produced by a virus encountered in studies of the 1933 St. Louis encephalitis epidemic. Public Health Reports (1896–1970). 1934;49(35):1019–27.
- Goldwater PN. A mouse zoonotic virus (LCMV): A possible candidate in the causation of SIDS. Med Hypotheses. 2021;158:
110735 . DOIPubMedGoogle Scholar - Ledesma J, Fedele CG, Carro F, Lledó L, Sánchez-Seco MP, Tenorio A, et al. Independent lineage of lymphocytic choriomeningitis virus in wood mice (Apodemus sylvaticus), Spain. Emerg Infect Dis. 2009;15:1677–80. DOIPubMedGoogle Scholar
- Ackermann R, Stille W, Blumenthal W, Helm EB, Keller K, Baldus O. [Syrian hamsters as vectors of lymphocytic choriomeningitis] [in German]. Dtsch Med Wochenschr. 1972;97:1725–31. DOIPubMedGoogle Scholar
- Ackermann R, Körver G, Turss R, Wönne R, Hochgesand P. [Prenatal infection with the virus of lymphocytic choriomeningitis: report of two cases (author’s transl)] [in German]. Dtsch Med Wochenschr. 1974;99:629–32. DOIPubMedGoogle Scholar
- Asper M, Hofmann P, Osmann C, Funk J, Metzger C, Bruns M, et al. First outbreak of callitrichid hepatitis in Germany: genetic characterization of the causative lymphocytic choriomeningitis virus strains. Virology. 2001;284:203–13. DOIPubMedGoogle Scholar
- Ackermann R, Stammler A, Armbruster B. Isolation of the lymphocytic choriomeningitis virus from curettage material after contact of the pregnant woman with a Syrian gold hamster (Mesocricetus auratus) [in German]. Infection. 1975;3:47–9. DOIGoogle Scholar
- Enders G, Varho-Göbel M, Löhler J, Terletskaia-Ladwig E, Eggers M. Congenital lymphocytic choriomeningitis virus infection: an underdiagnosed disease. Pediatr Infect Dis J. 1999;18:652–5. DOIPubMedGoogle Scholar
- Ackermann R, Bloedhorn H, Küpper B, Winkens I, Scheid W. [Spread of the lymphocytic choriomeningitis virus among West German mice. I. Investigations mostly on domestic mice (Mus musculus)] [in German]. Zentralbl Bakteriol Orig. 1964;194:407–30.PubMedGoogle Scholar
- Fornůsková A, Hiadlovská Z, Macholán M, Piálek J, de Bellocq JG. New perspective on the geographic distribution and evolution of lymphocytic choriomeningitis virus, central Europe. Emerg Infect Dis. 2021;27:2638–47. DOIPubMedGoogle Scholar
- Wasimuddin BJ, Bryja J, Ribas A, Baird SJ, Piálek J, Goüy de Bellocq J. Testing parasite ‘intimacy’: the whipworm Trichuris muris in the European house mouse hybrid zone. Ecol Evol. 2016;6:2688–701. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, et al. RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. DOIPubMedGoogle Scholar