Volume 29, Number 5—May 2023
Research Letter
Panton-Valentine Leukocidin–Positive CC398 MRSA in Urban Clinical Settings, the Netherlands
Figure
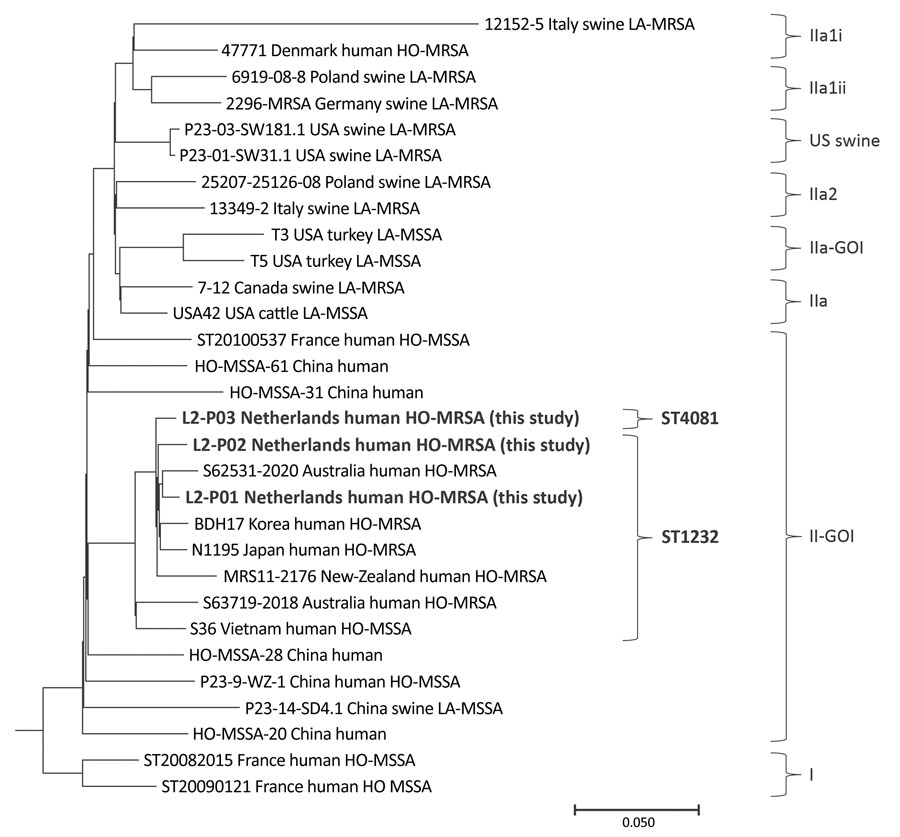
Figure. Maximum-parsimony tree of Panton-Valentine leukocidin–positive CC398 MRSA detected in urban clinical settings, the Netherlands. Bold text indicates 3 HO-MRSA strains detected in this study, which we deposited in GenBank (accession nos. SRR21673410 for L2-P01, SRR21624599 for L2-P02, and SRR21673965 for L2-P03); those strains were compared with 27 CC398 Staphylococcus aureus strains from GenBank. The tree was constructed by using GenBank accession no. AM990992.1 as a reference strain, and then rooted and grouped into clades, as previously described (1). Scale bar indicates nucleotide substitutions per 100 sites based on a concatenated alignment of 3,878 high-quality single-nucleotide polymorphisms. HO-MRSA, human-origin MRSA; LA-MRSA, livestock-associated MRSA; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-susceptible S. aureus; ST, sequence type.
References
- Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 2012;3:e00305–11. DOIPubMedGoogle Scholar
- Møller JK, Larsen AR, Østergaard C, Møller CH, Kristensen MA, Larsen J. International travel as source of a hospital outbreak with an unusual meticillin-resistant Staphylococcus aureus clonal complex 398, Denmark, 2016. Euro Surveill. 2019;24:
1800680 . DOIPubMedGoogle Scholar - Coombs GW, Daley D, Shoby P, Yee NWT, Robinson JO, Murray R, et al. Genomic characterisation of CC398 MRSA causing severe disease in Australia. Int J Antimicrob Agents. 2022;59:
106577 . DOIPubMedGoogle Scholar - Lu H, Zhao L, Si Y, Jian Y, Wang Y, Li T, et al. The surge of hypervirulent ST398 MRSA lineage with higher biofilm-forming ability is a critical threat to clinics. Front Microbiol. 2021;12:
636788 . DOIPubMedGoogle Scholar - Williamson DA, Bakker S, Coombs GW, Tan H, Monecke S, Heffernan H. Emergence and molecular characterization of clonal complex 398 (CC398) methicillin-resistant Staphylococcus aureus (MRSA) in New Zealand. J Antimicrob Chemother. 2014;69:1428–30. DOIPubMedGoogle Scholar
- Kaneko H, Kim ES, Yokomori S, Moon SM, Song KH, Jung J, et al. Comparative genomic analysis of the human variant of methicillin-resistant Staphylococcus aureus CC398 in Japan and Korea. Microb Drug Resist. 2022;28:330–7. DOIPubMedGoogle Scholar
- Konstantinovski MM, Schouls LM, Witteveen S, Claas ECJ, Kraakman ME, Kalpoe J, et al. Livestock-associated methicillin-resistant Staphylococcus aureus epidemiology, genetic diversity, and clinical characteristics in an urban region. Front Microbiol. 2022;13:
875775 . DOIPubMedGoogle Scholar - Strauß L, Ruffing U, Abdulla S, Alabi A, Akulenko R, Garrine M, et al. Detecting Staphylococcus aureus virulence and resistance genes: a comparison of whole-genome sequencing and DNA microarray technology. J Clin Microbiol. 2016;54:1008–16. DOIPubMedGoogle Scholar