Volume 29, Number 6—June 2023
Research Letter
Detection of Severe Murine Typhus by Nanopore Targeted Sequencing, China
Cite This Article
Citation for Media
Abstract
We report a case of murine typhus in China caused by Rickettsia typhi and diagnosed by nanopore targeted sequencing of a bronchoalveolar lavage fluid sample. This case highlights that nanopore targeted sequencing can effectively detect clinically unexplained infections and be especially useful for detecting infections in patients without typical signs and symptoms.
Murine typhus is caused by Rickettsia typhi bacteria transmitted by rat or cat flea vectors. Persons with murine typhus often have nonspecific or mild symptoms, such as fever, myalgia, and rash. In rare instances, murine typhus will cause atypical or multiple organ dysfunction syndrome (MODS) (1,2).
Murine typhus is an undifferentiated febrile illness, which makes it challenging to recognize and diagnose. We report a case of murine typhus and MODS in a patient without rash. We diagnosed murine typhus by using nanopore targeted sequencing (NTS) of a bronchoalveolar lavage fluid (BALF) sample, aiming to provide more reference for clinical practice.
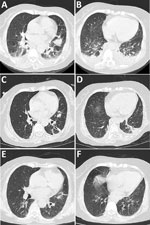
A 60-year-old female farmer from Yunnan Province, China, had fatigue, anorexia, nausea, dizziness, and vomiting for 1 week. At admission, she was afebrile and hemodynamically stable and did not have headache, rash, or eschar. Chest computed tomography (CT) imaging showed pneumonia and a small plural effusion (Figure, panels A, B). By the next day, her condition had deteriorated. She experienced chills, fever (temperature 39°C), severe hypotension (70/53 mm Hg), dyspnea, and deterioration of the oxygenation index. Preliminary laboratory investigation demonstrated mild leukocytosis (13.86 × 109 cells/L), moderately elevated transaminase levels (alanine aminotransferase 197 U/L, aspartate aminotransferase 128 U/L), severe thrombocytopenia (12 × 109 platelets/L), coagulation disorder (D-dimer 49.8 µg/mL), elevated C-reactive protein (207.4 mg/L) and procalcitonin (4.65 ng/mL) levels, and respiratory failure (partial pressure of oxygen 58.9 mm Hg).
The patient was given intravenous meropenem and norepinephrine and was admitted on noninvasive ventilation. We then conducted tests for malaria, Legionella, influenza virus, SARS-CoV-2, HIV, herpes simplex virus, cytomegalovirus, Epstein-Barr virus IgM, Roxiella burnettii IgM (phase II antigen), R. typhi IgM, Mycoplasma pneumoniae IgM, Chlamydia IgM, respiratory syncytial virus IgM, and adenovirus IgM; results were all negative. In addition, testing of blood, urine, stool, and sputum cultures and bone marrow biopsy all produced negative results.
On admission day 5, she remained normotensive. Her body temperature dropped, but she still had a low-grade fever, body temperature fluctuating from 37.5°C to 38°C. However, the cause of her severe infection remained unclear. The next day, she underwent bronchoscopy. BALF was sent to undergo NTS analysis to Wuhan Dgensee Clinical Laboratory Co., Ltd (https://www.dgensee.com). Two days later, NTS results revealed R. typhi DNA.
NTS analysis yielded a total of 71,252 single-end reads. R. typhi had the highest relative abundance of 44.58% (n = 31,764 reads). Subsequently, we conducted quantitative PCR (qPCR) using a sequence from GenBank (accession no. WP_011190964) as the target gene to detect R. typhi. The qPCR results confirmed the NTS detection (Appendix).
After the murine typhus diagnosis, the patient was treated with doxycycline (100 mg every 12 h) beginning on admission day 9. Soon after, her body temperature, platelet count, and blood coagulation function returned to normal. Reexamination of chest CT images showed improved pulmonary infiltrates (Figure, panels C, D), and she was discharged from the hospital. She continued doxycycline (100 mg 2×/d) for 14 days after discharge. At a 1-month follow-up, she had no symptoms of discomfort, and chest CT imaging showed resolving pulmonary infiltrates (Figure, panels E, F).
As an undifferentiated febrile illness, murine typhus can be challenging to diagnose in clinically mild or severe illness. Murine typhus can manifest with nonspecific symptoms and mimic other disease processes, making laboratory-confirmed diagnosis difficult without a high index of suspicion.
Diagnosis of murine typhus is usually performed by serologic testing and molecular analysis. Adaptation of modern serologic techniques for murine typhus diagnosis has substantially increased diagnostic accuracy, and serology was deemed the standard before the wide acceptance of molecular testing. Rickettsial infections require early diagnosis and treatment to prevent severe outcomes, but early diagnosis is rarely achieved by using serology (3). In addition to serology, the most common diagnostic method for murine typhus is qPCR (4). However, PCR has limitations, including being more sensitive during acute illness, such as the febrile phase, usually days 1–5 of illness, but possibly up to days 7–10 (3).
NTS is a groundbreaking technology that has the potential to overcome the shortcomings of both PCR and metagenomic next-generation sequencing, and next-generation sequencing is much less affected by antimicrobial drugs than is PCR (5–7). NTS has been used clinically and has shown high specificity and sensitivity (6). NTS combines long read length (>5,000 bp) and targeted amplification of 16S RNA gene for bacteria, rpoB for mycobacteria, and internal transcribed spacer (ITS) for fungi, all of which are free from interference of host background DNA (8,9). NTS can accurately detect causative pathogens in infectious samples and has a short 8–14-hour turnaround time.
In conclusion, we successfully detected R. typhi by using NTS in a febrile patient with MODS but without rash. NTS is a promising technology that can efficiently identify infectious pathogens early and has the potential to assist physicians in providing timely and precise treatment, especially for patients with nonspecific symptoms indicative of multiple disease processes.
Dr. Qian is a resident at the Affiliated Hospital of Yunnan University, Kunming, Yunnan, China. Her research interest is emerging infectious diseases.
Acknowledgments
We thank Dgensee, Wuhan, China for performing the targeted nanopore sequencing procedure and interpretation.
This work was supported by grants from the National Natural Science Foundation of China (grant no. 82160016), the Basic Research Program of Yunnan Province (grant no. 202201AY070001-265), and the Famous Doctors of High-Level Talent Training Support Program of Yunnan Province (grant no. YNWR-MY-2020-013).
References
- Civen R, Ngo V. Murine typhus: an unrecognized suburban vectorborne disease. Clin Infect Dis. 2008;46:913–8. DOIPubMedGoogle Scholar
- Tsioutis C, Chaliotis G, Kokkini S, Doukakis S, Tselentis Y, Psaroulaki A, et al. Murine typhus in elderly patients: a prospective study of 49 patients. Scand J Infect Dis. 2014;46:779–82. DOIPubMedGoogle Scholar
- Paris DH, Dumler JS. State of the art of diagnosis of rickettsial diseases: the use of blood specimens for diagnosis of scrub typhus, spotted fever group rickettsiosis, and murine typhus. Curr Opin Infect Dis. 2016;29:433–9. DOIPubMedGoogle Scholar
- Abdad MY, Abou Abdallah R, Fournier PE, Stenos J, Vasoo S. A concise review of the epidemiology and diagnostics of rickettsioses: Rickettsia and Orientia spp. J Clin Microbiol. 2018;56:e01728–17. DOIPubMedGoogle Scholar
- Hasan MR, Rawat A, Tang P, Jithesh PV, Thomas E, Tan R, et al. Depletion of human DNA in spiked clinical specimens for improvement of sensitivity of pathogen detection by next-generation sequencing. J Clin Microbiol. 2016;54:919–27. DOIPubMedGoogle Scholar
- Fu Y, Chen Q, Xiong M, Zhao J, Shen S, Chen L, et al. Clinical performance of nanopore targeted sequencing for diagnosing infectious diseases. Microbiol Spectr. 2022;10:
e0027022 . DOIPubMedGoogle Scholar - Ciuffreda L, Rodríguez-Pérez H, Flores C. Nanopore sequencing and its application to the study of microbial communities. Comput Struct Biotechnol J. 2021;19:1497–511. DOIPubMedGoogle Scholar
- Petersen LM, Martin IW, Moschetti WE, Kershaw CM, Tsongalis GJ. Third-generation sequencing in the clinical laboratory: exploring the advantages and challenges of nanopore sequencing. J Clin Microbiol. 2019;58:e01315–9. DOIPubMedGoogle Scholar
- Huang Q, Fu A, Wang Y, Zhang J, Zhao W, Cheng Y. Microbiological diagnosis of endophthalmitis using nanopore targeted sequencing. Clin Exp Ophthalmol. 2021;49:1060–8. DOIPubMedGoogle Scholar
Figure
Cite This ArticleOriginal Publication Date: May 08, 2023
Table of Contents – Volume 29, Number 6—June 2023
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Xiqian Xing, Department of Pulmonary and Critical Care Medicine, The Affiliated Hospital of Yunnan University, 176 Qingnian Rd, Kunming 650021, Yunnan, China
Top