Volume 29, Number 8—August 2023
Research
Genome-Based Epidemiologic Analysis of VIM/IMP Carbapenemase-Producing Enterobacter spp., Poland
Figure 1
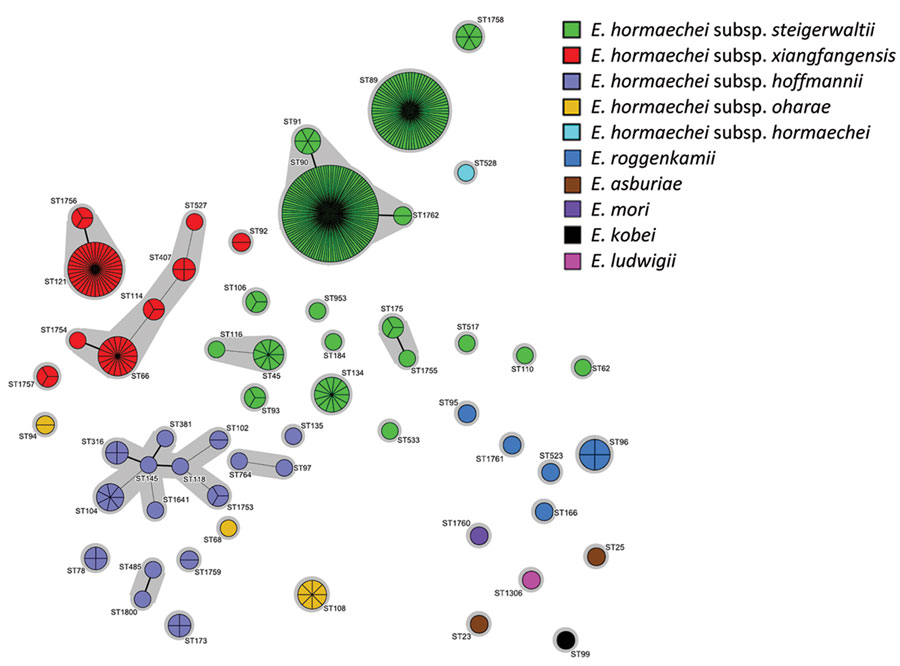
Figure 1. Population structure of Enterobacter spp. isolates identified in a genome-based epidemiologic analysis of VIM/IMP carbapenemase-producing Enterobacter spp., Poland, 2006–2019. The minimum-spanning tree was constructed on the basis of 7-loci multilocus sequence type data. Each circle represents 1 ST, and each fragment of a pie chart corresponds to 1 isolate. The size of a circle is proportional to the number of isolates of that ST. Connecting lines infer phylogenetic relatedness in terms of several allelic differences (thick solid line indicates a single-locus variant, thin solid line indicates a double-locus variant). ST, sequence type.
Page created: June 12, 2023
Page updated: July 20, 2023
Page reviewed: July 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.