Volume 29, Number 8—August 2023
Dispatch
Chromosome-Borne CTX-M-65 Extended-Spectrum β-Lactamase–Producing Salmonella enterica Serovar Infantis, Taiwan
Cite This Article
Citation for Media
Abstract
A CTX-M-65‒producing Salmonella enterica serovar Infantis clone, probably originating in Latin America and initially reported in the United States, has emerged in Taiwan. Chicken meat is the most likely primary carrier. Four of the 9 drug resistance genes have integrated into the chromosome: blaCTX-M-65, tet(A), sul1, and aadA1.
Salmonella enterica serovar Infantis is one of the most common Salmonella serotypes (1); it is frequently isolated from humans and animals, particularly from poultry (2). An increasing incidence of Salmonella Infantis infections has been reported in the United States (3), accompanied by emergence and spread of an extended-spectrum β-lactamase CTX-M-65‒producing Salmonella Infantis clone in humans, food animals, and retail chicken (4,5). The clone probably originated in South America because it was initially discovered in persons who had traveled back from Peru, Bolivia, Ecuador, and Chile since 2012 (5). Domestically acquired infections were not identified in the United States until 2014 (5).
This clone is characterized by having a D87Y mutation in the gyrA gene and carrying multiple resistance genes, including aph(4)-Ia, aac(3)-IVa, aph(3′)-Ic, blaCTX-M-65, fosA3, floR, dfrA14, sul1, tet(A), and aadA1, located in 2 distinct regions of a pESI-like megaplasmid (4). The CTX-M-65‒producing clone has been reported mostly in South America, North America, and some countries in Europe (4–12).
In Taiwan, Salmonella Infantis is not a common cause of human salmonellosis, accounting for only 0.61% (246/40,599) of all Salmonella isolates collected during 2004‒2022. Salmonella Infantis isolates collected during 2004‒2019 showed a low level of antimicrobial drug resistance (Appendix Table 1). However, in 2021, we identified that 7 of 14 Salmonella Infantis isolates from patients who had salmonellosis were multidrug-resistant (MDR), and in 2022, MDR strains accounted for 55% (21/38) of the Salmonella Infantis isolates recovered that year.
The 28 patients who contracted MDR Salmonella Infantis were from diverse age groups and geographic locations, and none of them had a history of international travel. During 2021 and 2022, the COVID-19 pandemic restricted travel abroad. We report a CTX-M-65‒producing Salmonella Infantis clone in Taiwan.
We performed clustering analysis on pulsed-field gel electrophoresis (PFGE) patterns of Salmonella Infantis isolates, which showed that the MDR isolates recovered in 2021 and 2022 clustered closely together in a distinct group (Appendix Figure). Antimicrobial drug susceptibility testing showed that the MDR isolates had resistance to ampicillin, cefotaxime, ceftazidime, nalidixic acid, ciprofloxacin (intermediate susceptibility), gentamicin, chloramphenicol, sulfamethoxazole, trimethoprim, and tetracycline (Appendix Figure). The resistance profile closely resembled that of the widespread CTX-M-65‒producing Salmonella Infantis clone (5).
We isolated Salmonella bacteria from retail raw chicken meat sold in 12 supermarket stores in Taichung City in 2022 to investigate the source of MDR Salmonella Infantis. All chicken meat samples were sourced from domestic farms. Salmonella bacteria were isolated from 191 (65.6%) of 291 chicken meat samples. A total of 379 Salmonella isolates were recovered from the 191 samples (1‒2 isolates from each Salmonella-positive sample).
Of the 379 isolates, 68.1% (258) were identified to be Salmonella Infantis, followed by Salmonella Kentucky (17.2%), Salmonella Brancaster (2.6%), Salmonella Goldcoast (2.6%), Salmonella Agona (2.4%), Salmonella Enteritidis (2.1%), and 6 other serovars (5.0%). Of the 191 samples, 11% were found to be contaminated with a mixture of Salmonella serovars. The 258 Salmonella Infantis isolates had 28 PFGE patterns, among which the 6 most common patterns were also observed in the MDR isolates from humans (Appendix Table 2). We performed a clustering analysis of PFGE profiles, which showed that the 258 Salmonella Infantis isolates from chicken meat, 28 MDR isolates from humans, and 1 isolate from a diseased pig recovered in 2022, were grouped in a common cluster (data not shown).
We conducted whole-genome sequencing of 51 Salmonella Infantis isolates from humans, chickens, and a pig by using the Illumina sequencing platform (https://www.illumina.com) to investigate drug resistance genetic determinants, plasmid incompatibility types, and their genetic relationships. Our analysis showed that all 51 Salmonella Infantis isolates belonged to sequence type 32, and 18 MDR Salmonella Infantis isolates recovered from humans, chickens, and a pig in 2021 and 2022 had a D87Y mutation in gyrA, along with an IncFIB plasmid and 4 common resistance genes: aadA1, blaCTX-M-65, sul1, and tet(A) (Appendix Table 3). In addition, 15 of the 18 blaCTX-M-65–carrying isolates had 5 other drug resistance genes: aac(3)-IVa, aph(3′)-Ia, aph(4)-Ia, dfrA14, and floR. Two of the isolates had 4 of the 5 drug resistance genes, and 1 did not have any of the 5 genes.
We conducted clustering analysis of core genome multilocus sequence typing profiles, which showed that the blaCTX-M-65–carrying isolates from Taiwan, when compared with non‒blaCTX-M-65–carrying strains, showed a closer genetic relationship with blaCTX-M-65–carrying strains reported in North and South America, Europe, Australia, India, and Vietnam (Figure 1).
To investigate the location of drug resistance genes, we performed additional sequencing of 6 blaCTX-M-65–carrying isolates and 1 pan-susceptible isolate by using the Oxford nanopore sequencing platform (https://nanoporetech.com). This approach provided long sequence reads, enabling us to assemble complete genome sequences. Our analysis showed that all 6 blaCTX-M-65–carrying isolates from humans, chickens, and a pig had 5 drug resistance genes, aac(3)-IVa, aph(3′)-Ia, aph(4)-Ia, dfrA14, and floR, within an ≈195-kb IncFIB plasmid. In contrast, aadA1, blaCTX-M-65, sul1, and tet(A) were found in an ≈126-kb DNA segment inserted within an ABC-F family ATPase gene in the chromosomes (Appendix Tables 4, 5).
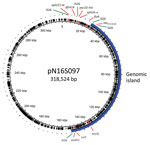
Our investigation suggested that the 195-kb IncFIB plasmids and the 126-kb genomic islands found in the chromosome probably originated from a plasmid similar to pN16S097. This megaplasmid, which has a length of 318,524 bp, was initially detected in a Salmonella Infantis strain and has 9 of the mentioned drug resistance genes in 2 distinct regions (8).
We hypothesize that the 126-kb segment carrying aadA1, blaCTX-M-65, sul1, and tet(A) might have translocated from a pN16S097-like plasmid into a chromosome through IS26-mediated transposition, resulting in formation of an 8-bp (CCGGAAAG) tandem repeat at the insertion site. This process led to the loss of the megaplasmid, leaving a plasmid of ≈195 kb (Figure 2). Upon analyzing 5,253 genomes of blaCTX-M-65–carrying Salmonella Infantis strains available in GenBank, we did not observe a large DNA segment or a blaCTX-M-65–carrying segment inserted within an ABC-F family ATPase gene in the chromosomes.
The blaCTX-M-65–carrying Salmonella Infantis clone, previously identified in South and North America and some countries in Europe, has been detected in Taiwan. Chickens are suspected to be the primary source of blaCTX-M-65–carrying strains. Many PFGE genotypes have been found among the isolates from retail chicken meat, indicating that the blaCTX-M-65–carrying Salmonella Infantis strains have probably evolved and proliferated on chicken farms, rather than being contaminants from chicken processing plants. Integration of blaCTX-M-65 into the chromosome suggests that this drug resistance gene might be more resiliently maintained within the strains.
Ms. Liao is a senior technical specialist at the Centers for Disease Control, Ministry of Health and Welfare, Taichung, Taiwan. Her primary research interests are molecular epidemiology and antimicrobial drug resistance of foodborne bacterial pathogens.
Acknowledgment
This study was supported by the Ministry of Health and Welfare, Taiwan (grant no. MOHW111-CDC-C-315-124306).
References
- Hendriksen RS, Vieira AR, Karlsmose S, Lo Fo Wong DM, Jensen AB, Wegener HC, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization Global Foodborne Infections Network Country Data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathog Dis. 2011;8:887–900. DOIPubMedGoogle Scholar
- European Food Safety A, European Centre for Disease P, Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022;20:
e07666 . - Collins JP, Shah HJ, Weller DL, Ray LC, Smith K, McGuire S, et al. Preliminary incidence and trends of infections caused by pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. Sites, 2016–2021. MMWR Morb Mortal Wkly Rep. 2022;71:1260–4. DOIPubMedGoogle Scholar
- Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, et al. Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother. 2017;61:e00488–17. DOIPubMedGoogle Scholar
- Brown AC, Chen JC, Watkins LKF, Campbell D, Folster JP, Tate H, et al. CTX-M-65 extended-spectrum β-lactamase-producing Salmonella enterica serotype Infantis, United States. Emerg Infect Dis. 2018;24:2284–91. DOIPubMedGoogle Scholar
- Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, et al. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One. 2015;10:
e0144802 . DOIPubMedGoogle Scholar - Hindermann D, Gopinath G, Chase H, Negrete F, Althaus D, Zurfluh K, et al. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front Microbiol. 2017;8:1322. DOIPubMedGoogle Scholar
- Tyson GH, Li C, Harrison LB, Martin G, Hsu CH, Tate H, et al. A multidrug-resistant Salmonella Infantis clone is spreading and recombining in the United States. Microb Drug Resist. 2021;27:792–9. DOIPubMedGoogle Scholar
- Bharat A, Mataseje L, Parmley EJ, Avery BP, Cox G, Carson CA, et al. One health genomic analysis of extended-spectrum β-lactamase‒producing Salmonella enterica, Canada, 2012‒2016. Emerg Infect Dis. 2022;28:1410–20. DOIPubMedGoogle Scholar
- Lee WWY, Mattock J, Greig DR, Langridge GC, Baker D, Bloomfield S, et al. Characterization of a pESI-like plasmid and analysis of multidrug-resistant Salmonella enterica Infantis isolates in England and Wales. Microb Genom. 2021;7:
000658 . DOIPubMedGoogle Scholar - Pietsch M, Simon S, Meinen A, Trost E, Banerji S, Pfeifer Y, et al. Third generation cephalosporin resistance in clinical non-typhoidal Salmonella enterica in Germany and emergence of blaCTX-M-harbouring pESI plasmids. Microb Genom. 2021;7:
000698 . DOIPubMedGoogle Scholar - Mejía L, Medina JL, Bayas R, Salazar CS, Villavicencio F, Zapata S, et al. Genomic epidemiology of Salmonella Infantis in Ecuador: from poultry farms to human infections. Front Vet Sci. 2020;7:
547891 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 29, Number 8—August 2023
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Chien-Shun Chiou, Central Region Laboratory, Center for Diagnostics and Vaccine Development, Centers for Disease Control, Taichung 40855, Taiwan
Top