Volume 29, Number 9—September 2023
Research
Genomic Characteristics of Emerging Intraerythrocytic Anaplasma capra and High Prevalence in Goats, China
Abstract
Anaplasma capra is an emerging tickborne human pathogen initially recognized in China in 2015; it has been reported in ticks and in a wide range of domestic and wild animals worldwide. We describe whole-genome sequences of 2 A. capra strains from metagenomic sequencing of purified erythrocytes from infected goats in China. The genome of A. capra was the smallest among members of the genus Anaplasma. The genomes of the 2 A. capra strains contained comparable G+C content and numbers of pseudogenes with intraerythrocytic Anaplasma species. The 2 A. capra strains had 54 unique genes. The prevalence of A. capra was high among goats in the 2 endemic areas. Phylogenetic analyses revealed that the A. capra strains detected in this study were basically classified into 2 subclusters with those previously detected in Asia. Our findings clarify details of the genomic characteristics of A. capra and shed light on its genetic diversity.
Anaplasma capra is an emerging tickborne zoonotic pathogen in the genus Anaplasma, family Anaplasmataceae, and was initially identified in blood samples from asymptomatic goats (Capra aegagrus hircus) and a febrile human patient with tick-bite history in China in 2015 (1). The patient infected with A. capra had fever, headache, malaise, dizziness, myalgia, gastrointestinal symptoms, rash, lymphadenopathy, and abnormalities in cerebrospinal fluid pleocytosis and hepatic aminotransferase. Since then, A. capra has been detected in various domestic animals (e.g., goats, sheeps, cattle, yaks, and dogs) (2–5) and wild animals (e.g., takins, muntjacs, water deer, musk deer, onagers, serows, and brown hares) (6–10), and in a wide range of ticks (e.g., Ixodes persulcatus, Haemaphysalis longicornis, H. qinghaiensis, Dermacentor abaensis, D. nuttalli, and Rhipicephalus microplus [1,11–14]) across China and around the world (2,7–10,15,16), posing a potential threat to the health of humans and animals.
Members of the family Anaplasmataceae have complex life cycles involving vertebrate hosts and hematophagous ticks, many of which have emerged as human pathogens. The genus Anaplasma was proposed according to the phylogenetic analyses based on 16S rRNA and groEL sequences (17) and initially encompassed 6 species: A. phagocytophilum, A. marginale, A. centrale, A. ovis, A. platys, and A. bovis. Subsequently, 2 candidate novel species (A. capra and A. odocoilei) and other unclassified genovariants (1,18–20) were included in the List of Prokaryotic Names with Standing in Nomenclature (https://www.bacterio.net) pending validation. To date, 5 Anaplasma species have been known to infect humans: A. phagocytophilum, A. capra, A. ovis, A. platys, and A. bovis (21). Since the A. marginale genome sequence was reported in 2005 (22), a total of 24 A. marginale genomes (23), 32 A. phagocytophilum genomes (24,25), 1 A. centrale genome (26), 2 A. ovis genomes (27), and 1 A. platys genome (28) have been sequenced and deposited in GenBank. Although A. capra has been extensively detected in ticks and animal hosts worldwide, no genome of the emerging pathogen has been determined so far, which has hindered us from better understanding its genetic features and pathogenesis. Considering A. capra is an intraerythrocytic pathogen and abundant in blood samples of host goats (1,29), we separated erythrocytes from the blood of infected goats to enrich the bacteria and generated the entire genome of A. capra using metagenome assembly to promote better understanding of this emerging pathogen, to compare the characteristics of A. capra genomes with previously published genomes of other Anaplasma and related species, and to evaluate intraspecies genetic diversity of A. capra in different geographic locations and tick species across China.
Sample Collection and Preparation
We collected EDTA blood samples from 3 flocks of goats in Shandong Province and a flock of goats in Guizhou Province, China (Appendix Figure 1), during September 2021–July 2022. Meanwhile, we prepared blood smears for some goats. We collected host-seeking ticks in the same areas where the infected goats lived by dragging white flags over vegetation. An entomologist (Y.S.) identified all ticks to the species level and developmental stage. We extracted DNA from each goat blood sample or tick by using a High Pure PCR Template Preparation Kit (Roche, https://www.roche.com) according to the manufacturer’s instructions.
PCRs and Sequencing
We conducted a nested PCR specific for the citrate synthase (gltA) gene of A. capra (Appendix Table 1) to screen all goat blood and tick samples, as previously described (1). We amplified all the positive samples for gltA by specific PCRs targeting the 16S rRNA, msp4, and groEL genes of A. capra (Appendix Table 1). We sequenced all amplicons to confirm the correctness of PCR results and conducted a SYBR Green–based quantitative PCR (qPCR) targeting different regions of the gltA gene by using a specific primer (Appendix Table 1).
Fluorescence In Situ Hybridization
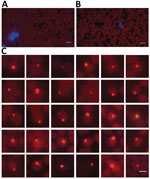
We used fluorescence in situ hybridization (FISH) to observe the A. capra on blood smears. We designed the probe on the basis of the 16S rRNA full-length sequence of A. capra (Appendix Table 2) and labeled it with Quasar 570. We resuspended the pooled FISH probes in a final concentration of 25 μmol/L in RNase-free storage buffer, which we protected from light and stored at –20°C. We performed FISH on the prepared blood smear with a commercial kit (Biosearch Technologies, https://www.biosearchtech.com), according to the manufacturer’s instructions.
Enrichment of A. capra for Genomic Sequencing
We separated erythrocytes from infected goats by conducting gradient centrifugation using cell separation solution (Eppendorf, https://www.eppendorf.com) for 20 min at 200 × g at 4°C. Then, we added 4 times volume of precooled (4°C) erythrocyte lysis buffer (Solarbio, http://www.solarbio.net) to the isolated erythrocytes by gentle pipetting to ensure adequate mixing. After placing the lysis solution at 4°C for 10 min, we centrifuged the solution at 350 × g for 10 min to remove residual blood cells. After that treatment, we maximally removed the host DNA in samples. Finally, we centrifuged the supernatant at 20,000 × g at 4°C for 30 min. We resuspended the pooled A. capra for DNA extraction by using the High Pure PCR Template Preparation Kit (Roche). We then constructed a sequencing library by using the AxyPrep MAG PCR Clean Up Kit (Fisher Scientific, https://www.fishersci.com) for an MGI sequencing set (https://en.mgi-tech.com). We prepared the sequencing library according to the Whole Genome Sequencing Library Preparation Protocol (MGI). We sequenced the paired-end libraries with a read length of 2 × 150 bp on a DNBseq-T7 platform at Grandomics Gene Technology Beijing Co. Ltd (Beijing, China).
Genome Assembly and Comparative Analyses
We mapped the clean reads to the goat (Capra hircus) reference genome (GenBank accession no. GCF_001704415) by using SAMtools 1.14 (30) to discard host-derived reads. We de novo assembled contigs from the unmapped reads by using metaSPAdes 3.15.3 (31). We performed contig binning by using MetaBAT 2.15 (32) and evaluated assembly quality by using CheckM version 1.1.3 in linage_wf mode, which searches for universal single-copy marker genes and deduces completeness and contamination on the basis of presence and absence of these genes (33). We generated G+C content, genome completeness, and annotation information and depicted them by using an approach described previously (34,35). We estimated average nucleotide identity (ANI) and DNA–DNA hybridization (DDH) by using fastANI 1.32 (36) and GGDC (https://ggdc.dsmz.de/ggdc.php).
Phylogenetic Analyses
We deposited in GenBank the results of the phylogenetic analysis of the whole genomes of the 2 A. capra strains and all the genomes of Anaplasma species by using Orthofinder 2.5.4 (37), after eliminating the poorly aligned positions and divergent regions by using Gblocks 0.91b. We aligned trimmed sequence by using Muscle 5.1 (R.C. Edgar, unpub. data, https://doi.org/10.1101/2021.06.20.449169) and constructed the phylogenetic tree by using iqtree 2.2.0.3 (38). Furthermore, we conducted phylogenetic analyses on A. capra gltA, groEL, 16S rRNA, and msp4 genes obtained from infected goats and ticks by using the maximum-likelihood method in MEGA11 (39).
Functional Analysis of Predicted Genes
To find difference in the Kyoto Encyclopedia of Genes and Genomes (KEGG) between the 2 strains of A. capra and other species in the genus Anaplasma, were annotated orthogroup sequences by using KOfam 1.4.0 (40) and illustrated them using a Venn diagram. We used the software eggNOG-Mapper 2.1.7 to determine the Clusters of Orthologous Group (COG) categories for protein encoding regions (41).
Forty-three (59.7%) of 72 goat blood samples were positive for gltA gene of A. capra. We chose 2 blood samples (1 from a 2-year-old female goat in Shandong Province and another from a 10-month-old female goat in Guizhou Province) (Appendix Figure 1) for next-generation sequencing because they had high bacterial loads (8.4 × 106 gltA gene copies/mL blood for the goat in Shandong Province and 2.0 × 106 gltA gene copies/mL blood for the goat in Guizhou Province) as estimated by qPCR (Appendix Table 1). In addition, we visualized A. capra by specific FISH in erythrocytes on the blood smear prepared from the goat in Shandong Province for next-generation sequencing (Figure 1).
The metagenome sequencing resulted in >38 million 150-bp clean reads from each sample. Despite primary removing of host DNA, 95.9% and 93.3% of reads in the 2 samples were mapped to the goat genome and discarded. The remaining reads were subsequently de novo assembled into contigs by using the SPAdes 3.15.3 with meta parameters (31). The 2 assembled A. capra genomes were named A. capra str. BIME1 (GenBank accession no. GCA_025628785.1) and A. capra str. BIME2 (GenBank accession no. GCA_025628805.1), and had a higher level of completeness (99.79% for BIME1 and 99.36% for BIME2). The genome of A. capra was the smallest (≈1.07 Mb) among those in the genus Anaplasma and the second smallest genome of the family Anaplasmataceae, just after Neorickettsia sennetsu (0.859 Mb) (24). The genome sequences of the 2 strains shared 99.89% nucleotide similarity with each other.
We compared the 2 A. capra genomes with other representative species strains in the genus Anaplasma (Appendix Table 3). The G+C content (48.3% for both) of the 2 A. capra genomes was similar to those of A. ovis, A. marginale, and A. centrale, which are all intraerythrocytic pathogens. The A. capra genomes yielded a total of 929 and 932 genes, of which 862 and 863, respectively, represented coding sequences. They possessed 37 tRNAs and a complete ribosomal RNA operon, in which the 16S rRNA gene was separated from the 23S-5S rRNA gene pair (Figure 2) as displayed by other members of the order Rickettsiales (42). The 2 strains of A. capra and other intraerythrocytic Anaplasma species, including A. ovis, A. centrale, and A. marginale, contained comparable numbers of pseudogenes that have lost functions owing to mutation accumulation and are observed more frequently in obligate intracellular bacteria where the lost gene functions are compensated by the host cells (43). Of note, A. phagocytophilum has ≈4-fold more pseudogenes than the other Anaplasma species (Appendix Table 3).
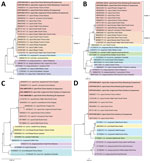
The estimated values of ANI and DDH between A. capra and other Anaplasma species suggested that A. capra were distinct from the other species. On the basis of ANI values, A. capra str. BIME1 was most similar to A. marginale, whereas A. capra str. BIME2 was most similar to A. ovis. The DDH results revealed that both A. capra strains were most close to A. marginale (Appendix Table 4). The phylogenetic analysis based on the single copy genes revealed that the 2 A. capra strains together occupied a distinct branch and were more closely related to A. ovis, A. marginale, and A. centrale than to A. phagocytophilum and A. platys in the genus Anaplasma (Figure 3, panel A). To explore the gene differences in species in the genus Anaplasma, we used Orthofinder (37) to identify the homologous genes. All species in the genus Anaplasma shared 643 genes in common, and the 2 A. capra strains together with other intraerythrocytic Anaplasma species (A. ovis, A. centrale, and A. marginale) shared 75 genes that are not present in the other 2 species, A. phagocytophilum and A. platys. Compared with other members of the genus Anaplasma, 14 genes were not possessed by A. capra. Of note, a total of 54 genes were only shared by the 2 A. capra strains, which had other 14 distinct genes in BIME1 and 10 in BIME2 (Figure 3, panel B). In addition, we identified 25 virulent genes in the 2 A. capra strains that were shared by all the species in the genus of Anaplasma, including virB2 gene family, virB6 gene family, virB4 gene family, virB8 gene family, virB9 gene family, and virB3, virB7, virB10, virB11, virD4, and Ats-1 genes that encode the type 4 secretion system and membrane protein-encoding genes (Appendix Table 5).
Among the 54 unique genes of A. capra, a total of 37 were unclassified, none of which was assigned to any KEGG category. Six of the remaining 17 genes were associated with metabolic processing, 5 genes were related to genetic information processing, and 6 were involved in signaling and cellular processing (Appendix Table 6). Among them, the most noteworthy of genes were RSF1, a gene related to the repair of DNA double-strand breaks (44), and desk, which encodes a protein acting as a kinase at cold temperatures in Bacillus subtilis (45).
We classified the coding proteins of the 2 A. capra strains (BIME1 and BIME2) into functional clusters of orthologous group (COG) categories and compared them with those of representative species strains in the genus Anaplasma (Appendix Table 7). Most proteins were involved in translation, ribosomal structure and biogenesis, energy production and conversion, and nutrient (including amino acid, nucleotide, carbohydrate, coenzyme, and lipid) transport and metabolism, all of which were essential for bacterial survival. Of note, the number of genes encoding cell wall and membrane in A. platys was substantially lower than those of other Anaplasma species. In addition, ≈10% of the proteins did not assign to any COG category and were classified as function unknown in each species.
We screened blood samples from 3 flocks of 54 goats in Shandong Province and a flock of 18 goats in Guizhou Province (Appendix Figure 1) by using nested PCR and qPCR targeting different regions of the gltA gene (Appendix Table 1). The overall positive rate was 59.7% (95% CI 48.4%–71.0%), and the positive rate was significantly higher among goats in Guizhou Province than in Shandong Province (77.8% vs. 53.7%; p<0.001). Accordingly, among the H. longicornis ticks collected from the same sites of the positive goats, the overall positive rate was 8.0% (95% CI 4.2%–11.8%), and the A. capra infection rate was significantly higher among ticks in Guizhou Province than that in Shandong Province (15.8% vs. 4.9%; p<0.001) (Appendix Table 8). To understand the genetic diversity, we amplified A. capra 16S rRNA (1,500 bp), groEL (1264 bp), and msp4 (799 bp) genes from those positive samples. We compared the nucleotide identities for each gene sequence and (Appendix Figures 2–5; GenBank accession numbers are provided).
The gltA genes amplified from either goats or ticks in this study had 99.7%–100% identity with each other and with the strain that infected humans (Appendix Figure 2). The phylogenetic analysis based on gltA gene revealed that the A. capra sequences in this study were in an independent cluster from those previously reported in various animals from China and South Korea but distinct from those detected in wild and domestic animals from Europe and Kyrgystan. The South Korea water deer seemed to be capable of carrying both variants of A. capra (Figure 4, panel A). No A. capra groEL gene was acquired from tick samples, and the sequences from goats shared 99.4%–100% identity with each other and 99.8%–100% with sequences from humans (Appendix Figure 3). Similarly, the phylogenetic analyses based on the groEL gene revealed that A. capra strains of this study clustered with those from humans, dogs, and domestic ruminants in Asia but were distinguished from those in Europe (Figure 4, panel B). The entire 16S rRNA gene sequences (1,500 bp) of A. capra detected in goats and H. longicornis ticks from either Shandong or Guizhou Province shared average similarity of >99.7% from each other and from the sequence detected in humans (Appendix Figure 4). The phylogenetic tree based on 16S rRNA gene sequences indicated that all the A. capra strains detected in this study were in the same clade with previously reported strains in Asia (Figure 4, panel C). The A. capra msp4 gene sequences were also relatively conserved (Appendix Figure 5) among the goats and ticks, and the topology of phylogenetic tree based on msp4 gene were similar to that based on the 16S rRNA gene, in which all A. capra sequences clustered in the clade different from other members of Anaplasma species (Figure 4, panel D).
Whole-genome assembly of obligate intracellular bacteria has usually been hindered by the DNA presence of host cells. In this study, we first assembled 2 complete genomes of A. capra from the red blood cells of infected goats by using the metagenomic sequencing strategy. Because A. capra is an intraerythrocytic pathogen (1,29), we separated erythrocytes from the periphery blood of the infected goats and then lysed them for maximum removal of goat DNA. After metagenomic next-generation sequencing, we discarded the remaining goat genomic sequences and successfully assembled the A. capra genomes from 2 infected goats. The high percentage of reads from goat could be attributable to the low abundance of A. capra in erythrocytes or the fact that all other host cells rather than erythrocytes were not totally removed during the isolation of erythrocytes. In any case, the completeness of the 2 A. capra genomes are up to 99.79% for BIME1 and 99.36% for BIME2. The genome sizes obtained in this study reach 1,066,874 bp for BIME1 and 1,059,758 bp for BIME2. Therefore, their predicted sizes are ≈1.07 Mbp, which remain the smallest genome in the genus of Anaplasma. The phylogenetic analysis based on genome sequences and the comparative analyses of genomic characteristics provide the evidence that A. capra is closely related to other intraerythrocytic Anaplasma species, including A. ovis, A. centrale, and A. marginale.
The genome of A. capra consists of a single circular chromosome with a total size of 1.07 Mbp and has 862 protein-coding genes, which is smaller than other Anaplasma species. In fact, all the Anaplasma genomes sequenced so far are relatively small compared with free-living bacteria. The small genome size might be because a part of the intracellular bacterial functions has been compensated by the host cells, a process of reductive evolution that has occurred in the order Rickettsiales because of long-term intracellular association with eukaryotic hosts (46). This reductive evolution is associated with the frequent formation of pseudogenes, affecting distinct loci in different species (47). Moreover, we found that the G+C content of A. capra is close to that of A. ovis, A. marginale, and A. centrale. Of note, their relatedness also seems to be closest according to the phylogenetic analysis. The common invasiveness of erythrocytes also accounts for their high similarity.
A limitation of this study is that both the A. capra genomes were directly derived from the blood samples of infected goats through metagenomic next-generation sequencing. Unfortunately, we did not obtain the genomes at chromosome level, which usually relies on 3rd-generation sequencing of an isolate. In any case, this study reveals the genomic characteristics of A. capra and sheds light on its genetic diversity.
The high prevalence of A. capra in goats from Shandong and Guizhou Provinces in this study further indicate that domestic ruminants might be the main animal hosts, as suggested by previous studies (2–5). H. longicornis ticks collected from the same sites of the positive goats either in Shandong Province or Guizhou Province are naturally infected with A. capra, implying the role of the tick species in transmission of the pathogen. Phylogenetic analyses based on the gltA and groEL genes demonstrate that A. capra strains detected from goats and H. longicornis ticks in this study are clustered in the same clade with those from humans, domestic ruminants, dogs, and Korean water deer (2,3,5,10). Of note, another clade of A. capra strains is mainly found in the wild and domestic animals from Europe and Kyrgyzstan (6,10,48). Those findings suggest that the enzootic cycles in various regions of the world might be different. Public health professionals should pay enough attention and formulate prevention and control strategies to reduce the health threat of the emerging tickborne pathogen to humans in other countries besides China.
Mr. Lin is an MPH candidate in the State Key Laboratory of Pathogen and Biosecurity at the Beijing Institute of Microbiology and Epidemiology. His primary research interests are the prevention and control of tickborne pathogens.
Acknowledgment
This study was supported by the Natural Science Foundation of China (grant no. 81621005 to W.-C.C.; grant nos. 81760605, 82160633, and 82103897 to L.Z.), the Natural Science Foundation of Shandong Province, China (grant no. ZR2020QH299 to L.Z.), and the Cheeloo Young Scholar Program of Shandong University.
References
- Li H, Zheng YC, Ma L, Jia N, Jiang BG, Jiang RR, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15:663–70. DOIPubMedGoogle Scholar
- Yang J, Liu Z, Niu Q, Liu J, Han R, Guan G, et al. A novel zoonotic Anaplasma species is prevalent in small ruminants: potential public health implications. Parasit Vectors. 2017;10:264. DOIPubMedGoogle Scholar
- Seo MG, Ouh IO, Lee H, Geraldino PJL, Rhee MH, Kwon OD, et al. Differential identification of Anaplasma in cattle and potential of cattle to serve as reservoirs of Anaplasma capra, an emerging tick-borne zoonotic pathogen. Vet Microbiol. 2018;226:15–22. DOIPubMedGoogle Scholar
- Wang Y, Zhang Q, Han S, Li Y, Wang B, Yuan G, et al. Ehrlichia chaffeensis and four Anaplasma species with veterinary and public health significance identified in Tibetan sheep (Ovis aries) and yaks (Bos grunniens) in Qinghai, China. Front Vet Sci. 2021;8:
727166 . DOIPubMedGoogle Scholar - Shi K, Li J, Yan Y, Chen Q, Wang K, Zhou Y, et al. Dogs as new hosts for the emerging zoonotic pathogen Anaplasma capra in China. Front Cell Infect Microbiol. 2019;9:394. DOIPubMedGoogle Scholar
- Yang J, Liu Z, Niu Q, Mukhtar MU, Guan G, Liu G, et al. A novel genotype of “Anaplasma capra” in wildlife and its phylogenetic relationship with the human genotypes. Emerg Microbes Infect. 2018;7:210. DOIPubMedGoogle Scholar
- Staji H, Yousefi M, Hamedani MA, Tamai IA, Khaligh SG. Genetic characterization and phylogenetic of Anaplasma capra in Persian onagers (Equus hemionus onager). Vet Microbiol. 2021;261:
109199 . DOIPubMedGoogle Scholar - Rocchigiani G, Ebani VV, Nardoni S, Bertelloni F, Bascherini A, Leoni A, et al. Molecular survey on the occurrence of arthropod-borne pathogens in wild brown hares (Lepus europaeus) from Central Italy. Infect Genet Evol. 2018;59:142–7. DOIPubMedGoogle Scholar
- Sato M, Nishizawa I, Fujihara M, Nishimura T, Matsubara K, Harasawa R. Phylogenetic analysis of the 16S rRNA gene of Anaplasma species detected from Japanese serows (Capricornis crispus). J Vet Med Sci. 2009;71:1677–9. DOIPubMedGoogle Scholar
- Amer S, Kim S, Yun Y, Na KJ. Novel variants of the newly emerged Anaplasma capra from Korean water deer (Hydropotes inermis argyropus) in South Korea. Parasit Vectors. 2019;12:365. DOIPubMedGoogle Scholar
- Yang J, Liu Z, Niu Q, Liu J, Han R, Liu G, et al. Molecular survey and characterization of a novel Anaplasma species closely related to Anaplasma capra in ticks, northwestern China. Parasit Vectors. 2016;9:603. DOIPubMedGoogle Scholar
- Zhang H, Sun Y, Jiang H, Huo X. Prevalence of severe febrile and thrombocytopenic syndrome virus, Anaplasma spp. and Babesia microti in hard ticks (Acari: Ixodidae) from Jiaodong Peninsula, Shandong Province. Vector Borne Zoonotic Dis. 2017;17:134–40. DOIPubMedGoogle Scholar
- Han R, Yang JF, Mukhtar MU, Chen Z, Niu QL, Lin YQ, et al. Molecular detection of Anaplasma infections in ixodid ticks from the Qinghai-Tibet Plateau. Infect Dis Poverty. 2019;8:12. DOIPubMedGoogle Scholar
- Guo WP, Zhang B, Wang YH, Xu G, Wang X, Ni X, et al. Molecular identification and characterization of Anaplasma capra and Anaplasma platys-like in Rhipicephalus microplus in Ankang, Northwest China. BMC Infect Dis. 2019;19:434. DOIPubMedGoogle Scholar
- Remesar S, Prieto A, García-Dios D, López-Lorenzo G, Martínez-Calabuig N, Díaz-Cao JM, et al. Diversity of Anaplasma species and importance of mixed infections in roe deer from Spain. Transbound Emerg Dis. 2022;69:e374–85. DOIPubMedGoogle Scholar
- Elhachimi L, Rogiers C, Casaert S, Fellahi S, Van Leeuwen T, Dermauw W, et al. Ticks and tick-borne pathogens abound in the cattle population of the Rabat-Sale Kenitra Region, Morocco. Pathogens. 2021;10:1594. DOIPubMedGoogle Scholar
- Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–65. DOIPubMedGoogle Scholar
- Tate CM, Howerth EW, Mead DG, Dugan VG, Luttrell MP, Sahora AI, et al. Anaplasma odocoilei sp. nov. (family Anaplasmataceae) from white-tailed deer (Odocoileus virginianus). Ticks Tick Borne Dis. 2013;4:110–9. DOIPubMedGoogle Scholar
- Calchi AC, Vultão JG, Alves MH, Yogui DR, Desbiez ALJ, De Santi M, et al. Ehrlichia spp. and Anaplasma spp. in Xenarthra mammals from Brazil, with evidence of novel ‘Candidatus Anaplasma spp.’. Sci Rep. 2020;10:12615. DOIPubMedGoogle Scholar
- Vanstreels RET, Yabsley MJ, Parsons NJ, Swanepoel L, Pistorius PA. A novel candidate species of Anaplasma that infects avian erythrocytes. Parasit Vectors. 2018;11:525. DOIPubMedGoogle Scholar
- Rar V, Tkachev S, Tikunova N. Genetic diversity of Anaplasma bacteria: Twenty years later. Infect Genet Evol. 2021;91:
104833 . DOIPubMedGoogle Scholar - Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, et al. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci U S A. 2005;102:844–9. DOIPubMedGoogle Scholar
- Dall’Agnol B, Webster A, Souza UA, Barbieri A, Mayer FQ, Cardoso GA, et al. Genomic analysis on Brazilian strains of Anaplasma marginale. Rev Bras Parasitol Vet. 2021;30:
e000421 . DOIPubMedGoogle Scholar - Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, et al. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:
e21 . DOIPubMedGoogle Scholar - Barbet AF, Al-Khedery B, Stuen S, Granquist EG, Felsheim RF, Munderloh UG. An emerging tick-borne disease of humans is caused by a subset of strains with conserved genome structure. Pathogens. 2013;2:544–55. DOIPubMedGoogle Scholar
- Herndon DR, Palmer GH, Shkap V, Knowles DP Jr, Brayton KA. Complete genome sequence of Anaplasma marginale subsp. centrale. J Bacteriol. 2010;192:379–80. DOIPubMedGoogle Scholar
- Liu Z, Peasley AM, Yang J, Li Y, Guan G, Luo J, et al. The Anaplasma ovis genome reveals a high proportion of pseudogenes. BMC Genomics. 2019;20:69. DOIPubMedGoogle Scholar
- Llanes A, Rajeev S. First whole genome sequence of Anaplasma platys, an obligate intracellular rickettsial pathogen of dogs. Pathogens. 2020;9:277. DOIPubMedGoogle Scholar
- Peng Y, Lu C, Yan Y, Song J, Pei Z, Gong P, et al. The novel zoonotic pathogen, Anaplasma capra, infects human erythrocytes, HL-60, and TF-1 cells in vitro. Pathogens. 2021;10:600. DOIPubMedGoogle Scholar
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience. 2021;10:giab008.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. metaSPAdes: a new versatile metagenomic assembler. Genome Res. 2017;27:824–34. DOIPubMedGoogle Scholar
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinformatics. 2020;70:
e102 . DOIPubMedGoogle Scholar - Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Syberg-Olsen MJ, Garber AI, Keeling PJ, McCutcheon JP, Husnik F. Pseudofinder: detection of pseudogenes in prokaryotic genomes. Mol Biol Evol. 2022;39:msac153.
- Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9:5114. DOIPubMedGoogle Scholar
- Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38:3022–7. DOIPubMedGoogle Scholar
- Aramaki T, Blanc-Mathieu R, Endo H, Ohkubo K, Kanehisa M, Goto S, et al. KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics. 2020;36:2251–2. DOIPubMedGoogle Scholar
- Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol Biol Evol. 2021;38:5825–9. DOIPubMedGoogle Scholar
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Pontén T, Alsmark UC, Podowski RM, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–40. DOIPubMedGoogle Scholar
- Beare PA, Sandoz KM, Omsland A, Rockey DD, Heinzen RA. Advances in genetic manipulation of obligate intracellular bacterial pathogens. Front Microbiol. 2011;2:97. DOIPubMedGoogle Scholar
- Helfricht A, Wiegant WW, Thijssen PE, Vertegaal AC, Luijsterburg MS, van Attikum H. Remodeling and spacing factor 1 (RSF1) deposits centromere proteins at DNA double-strand breaks to promote non-homologous end-joining. Cell Cycle. 2013;12:3070–82. DOIPubMedGoogle Scholar
- Albanesi D, Martín M, Trajtenberg F, Mansilla MC, Haouz A, Alzari PM, et al. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci U S A. 2009;106:16185–90. DOIPubMedGoogle Scholar
- Darby AC, Cho NH, Fuxelius HH, Westberg J, Andersson SG. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 2007;23:511–20. DOIPubMedGoogle Scholar
- Ogata H, Audic S, Renesto-Audiffren P, Fournier PE, Barbe V, Samson D, et al. Mechanisms of evolution in Rickettsia conorii and R. prowazekii. Science. 2001;293:2093–8. DOIPubMedGoogle Scholar
- Jouglin M, Rispe C, Grech-Angelini S, Gallois M, Malandrin L. Anaplasma capra in sheep and goats on Corsica Island, France: A European lineage within A. capra clade II? Ticks Tick Borne Dis. 2022;13:
101934 . DOIPubMedGoogle Scholar
Figures
Cite This ArticleOriginal Publication Date: August 08, 2023
1These senior authors contributed equally to this article.
Table of Contents – Volume 29, Number 9—September 2023
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Wu-Chun Cao, State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, 20 Dong-Da St, Fengtai District, Beijing 100071, China
Top