Volume 30, Number 1—January 2024
Research
Disease-Associated Streptococcus pneumoniae Genetic Variation
Figure 3
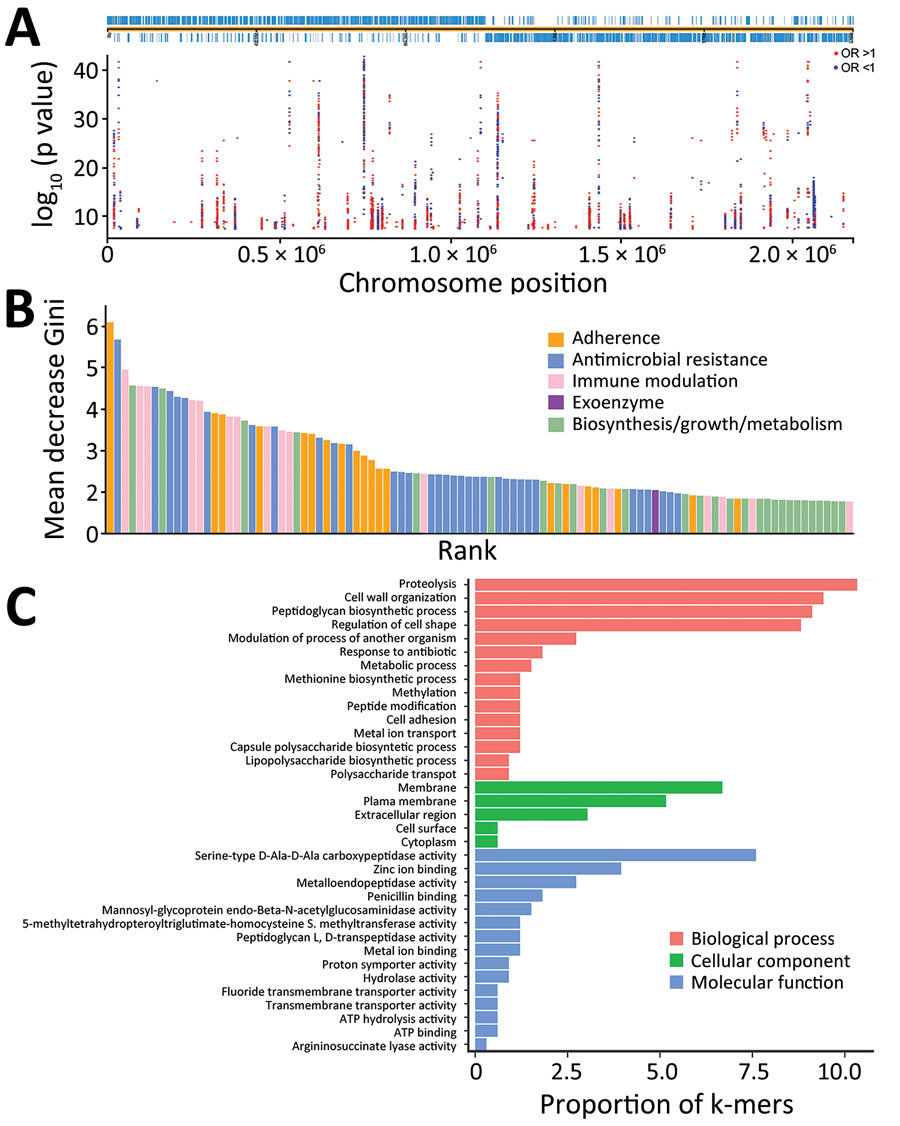
Figure 3. Preliminary screening for infection-associated k-mers by linear mixed model in study of disease-associated Streptococcus pneumoniae genetic variation. A) Manhattan plot showing statistical significance and chromosomal location of k-mers mapped to a complete reference genome (TIGR4; GenBank accession no. NC_003028.3). B) Importance of the top 100 k-mer predictors in a simpler model with 886 k-mers. C) Gene ontology annotations of the top 100 k-mer predictors. OR, odds ratio.
1These authors contributed equally to this article.
Page created: November 24, 2023
Page updated: December 20, 2023
Page reviewed: December 20, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.