Volume 30, Number 10—October 2024
Research Letter
Fort Sherman Virus Infection in Human, Peru, 2020
Figure 2
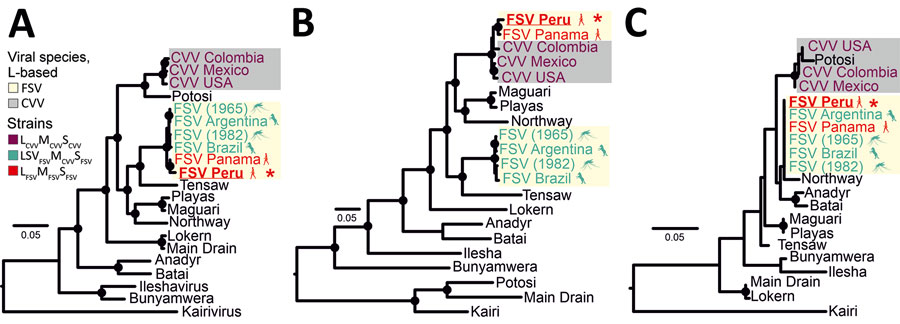
Figure 2. Maximum-likelihood phylogenetic trees based on deduced amino acid sequences of the L (A), M (B), and S (C) gene segments in study of Fort Sherman virus infection, Peru, 2020. Red asterisks indicate the FSV strain sequenced in this study. Black circles at nodes represent support values of >0.70 from 1,000 bootstrap replicates. Additional information on the reference sequences used to build the tree is provided (Appendix Table 1). Scale bars indicate genetic distance. CVV, Cache Valley virus; FSV, Fort Sherman virus; L, large segment; M, medium segment; S, small segment.
Page created: September 04, 2024
Page updated: September 24, 2024
Page reviewed: September 24, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.