Volume 30, Number 10—October 2024
Dispatch
Dengue Virus Serotype 3 Origins and Genetic Dynamics, Jamaica
Figure 3
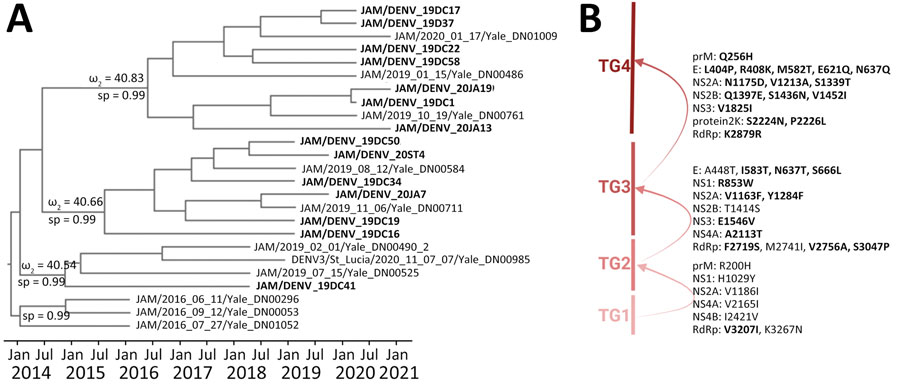
Figure 3. Time-scaled phylogenetic analysis, molecular characterization, dynamics, and natural selection of dengue virus serotype 3 in Jamaica. A) Phylogenetic tree indicates monophyletic clusters of strains from Jamaica (bold text) extracted from the discrete phylogeographic analysis (Figure 2). Discrete sp values (ω) for nodes evaluated for episodic selection are shown. Full sp values for nodes are shown in Appendix Figure 2, panel B. B) Strains were evaluated for amino acid replacements according to each TG. Arrows indicate episodic selection of each main TG clade. Bold text indicates positively selected mutations. sp, state probability; TG, temporal group.
1These first authors contributed equally to this article.
Page created: August 01, 2024
Page updated: September 23, 2024
Page reviewed: September 23, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.