Volume 30, Number 10—October 2024
Dispatch
Oxacillinase-484–Producing Enterobacterales, France, 2018–2023
Figure 2
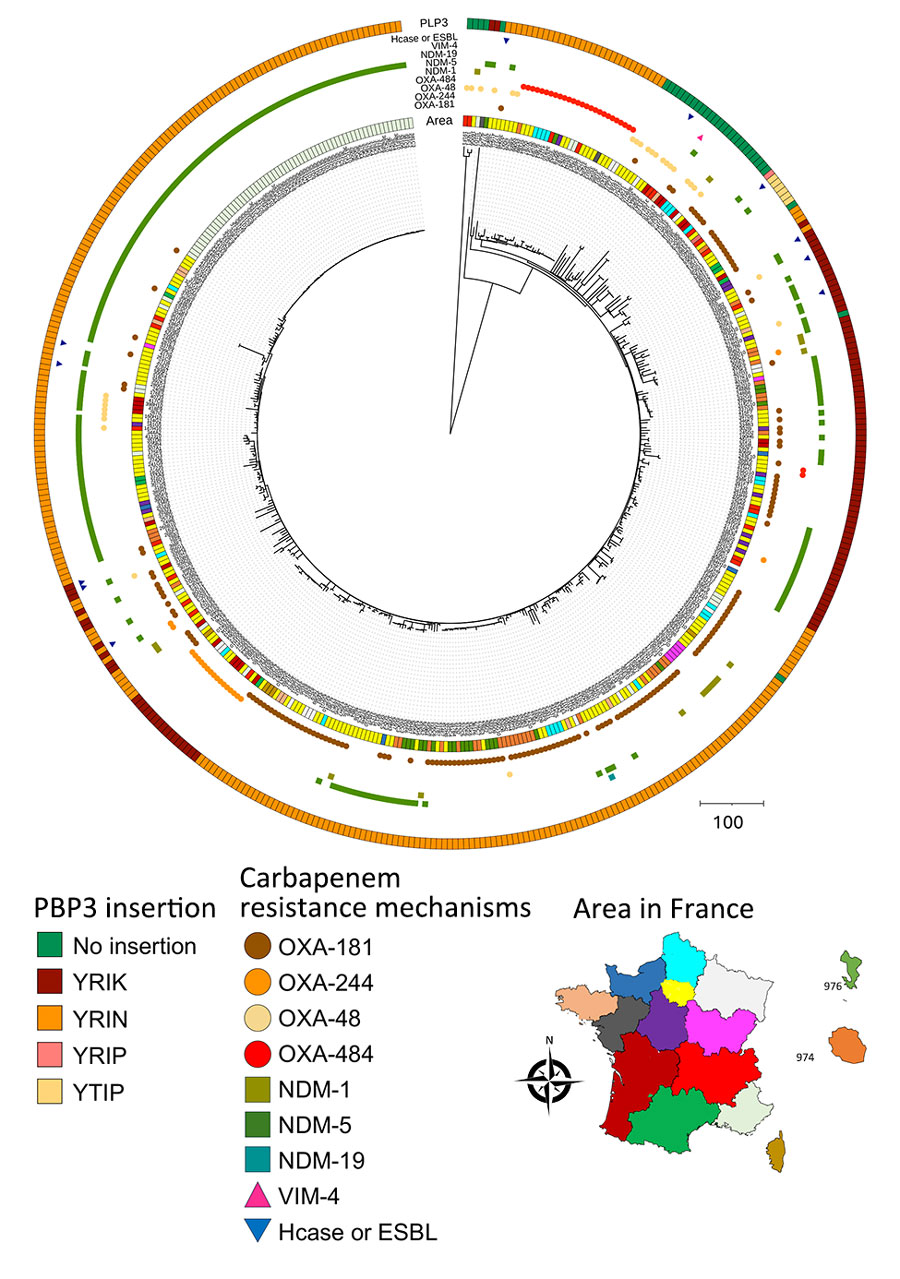
Figure 2. Phylogenetic tree and location of carbapenem-resistant Escherichia coli sequence type (ST) 410 isolates received at the French National Reference Center for Carbapenem-Resistant Enterobacterales, France, 2012–2023. Carbapenemases are classified into 3 of the 4 Ambler classes: class A (mainly Klebsiella pneumoniae carbapenemase); class B or metallo-β-lactamases (predominantly NDM, VIM, or imipenemase types); and class D, primarily OXA-48–like types, including OXA-484 producers. Carbapenemase types are detailed and OXA-484 producers are indicated. ST410 strains are distinguished by whether there are amino acid insertions (YRIK, YRIN, YRIP, or YTIP motifs) in PBP3. We conducted the single-nucleotide polymorphism analysis on a common genome representing 89.05% of the entire genome of the reference strain 303D1. Map shows locations where specific mechanisms were found. ESBL, extended-spectrum β-lactamase; Hcase, helicase; NDM, New Delhi metallo-β-lactamase; OXA, oxacillinase; PBP3, penicillin-binding protein 3; VIM, Verona integron-encoded metallo-β-lactamase.