Presumed Transmission of 2 Distinct Monkeypox Virus Variants from Central African Republic to Democratic Republic of the Congo
Emmanuel Hasivirwe Vakaniaki, Eddy Kinganda-Lusamaki, Sydney Merritt, Francois Kasongo, Emile Malembi, Lygie Lunyanga, Sylvie Linsuke, Megan Halbrook, Ernest Kalthan, Elisabeth Pukuta, Adrienne Amuri Aziza, Jean Claude Makangara Cigolo, Raphael Lumembe, Gabriel Kabamba, Yvon Anta, Pierrot Bolunza, Innocent Kanda, Raoul Ngazobo, Thierry Kalonji, Justus Nsio, Patricia Matoka, Dieudonné Mwamba, Christian Ngandu, Souradet Y. Shaw, Robert Shongo, Joule Madinga, Yap Boum, Laurens Liesenborghs, Eric Delaporte, Ahidjo Ayouba, Nicola Low, Steve Ahuka Mundeke, Lisa E. Hensley, Jean-Jacques Muyembe Tamfum, Emmanuel Nakoune, Martine Peeters, Nicole A. Hoff, Jason Kindrachuk, Anne W. Rimoin
1
, and Placide Mbala-Kingebeni
1
Author affiliations: Institut National de Recherche Biomédicale, Kinshasa, Democratic Republic of the Congo (E.H. Vakaniaki, E. Kinganda-Lusamaki, F. Kasongo, L. Lunyanga, S. Linsuke, E. Pukuta, A.A. Aziza, J.C. Makangara Cigolo, R. Lumembe, G. Kabamba, Y. Anta, J. Madinga, S.A. Mundeke, J.-J. Muyembe Tamfum, P. Mbala-Kingebeni); Institute of Tropical Medicine, Antwerp, Belgium (E.H. Vakaniaki, L. Liesenborghs); Cliniques Universitaires de Kinshasa, Kinshasa University, Kinshasa (E. Kinganda-Lusamaki, J.C. Makangara Cigolo, R. Lumembe, G. Kabamba, S.A. Mundeke, J.-J. Muyembe Tamfum, P. Mbala-Kingebeni); Université de Montpellier, French National Research Institute for Sustainable Development, INSERM, Montpellier, France (E. Kinganda-Lusamaki, E. Delaporte, A. Ayouba, M. Peeters); University of California, Los Angeles, California, USA (S. Merritt, M. Halbrook, N.A. Hoff, A.W. Rimoin); Hemorrhagic Fevers and Monkeypox Program, Ministry of Health, Kinshasa (E. Malembi, T. Kalonji, R. Shongo); Ministry of Health and Populations, Bangui, Central African Republic (E. Kalthan); University of Bern, Bern, Switzerland (J.C. Makangara Cigolo, N. Low); Provincial Health Division, South Ubangi, Democratic Republic of the Congo (P. Bolunza, I. Kanda); Mbaya Health Zone, South Ubangi (R. Ngazobo); National Border Hygiene Program, Ministry of Health, Kinshasa (J. Nsio, P. Matoka); National Institute of Public Health, Ministry of Health, Kinshasa (D. Mwamba, C. Ngandu); University of Manitoba, Winnipeg, Manitoba, Canada (S.Y. Shaw, J. Kindrachuk); Pasteur Institute of Bangui, Bangui, Central African Republic (Y. Boum, E. Nakoune); USDA Agricultural Research Service, Manhattan, Kansas, USA (L.E. Hensley)
Main Article
Figure 4
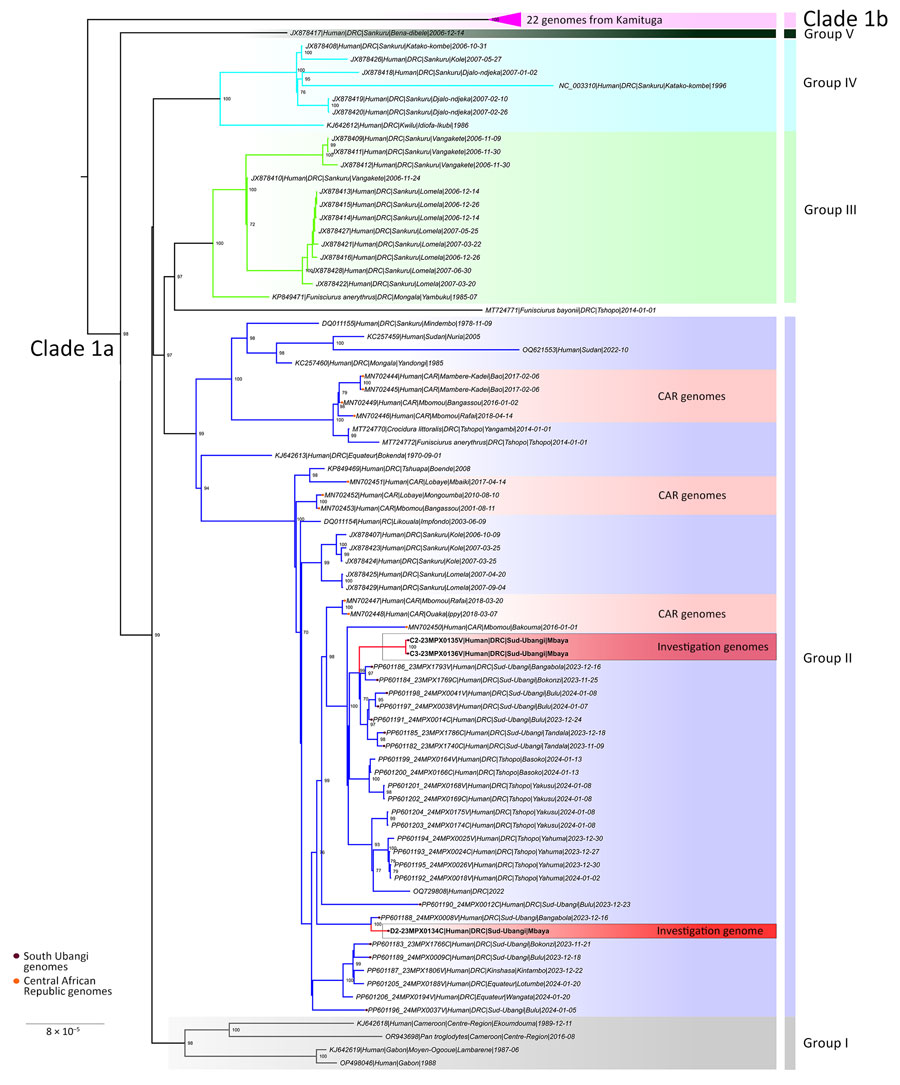
Figure 4. Phylogenetic analysis of monkeypox virus (MPXV) sequences from Mbaya Health Zone in study of presumed transmission of MPXV variants from Central African Republic to Democratic Republic of the Congo. Phylogenetic analysis of MPXV genome sequences are from samples described in this study and clade I MPXV sequences from Central Africa. Bootstrap support values are shown at branch points. DNA was extracted at the National Institute for Biomedical Research using a QIAGEN DNA Mini Kit (https://www.qiagen.com) from blood samples and subsequently screened for MPXV with an orthopoxvirus-specific real-time PCR assay. Whole-genome sequencing was attempted on samples from the index case by next-generation sequencing. Library preparation was performed using Illumina DNA Prep with Enrichment (https://www.illumina.com), and libraries were enriched for MPXV using biotinylated custom probes synthesized by Twist Biosciences (https://www.twistbioscience.com). Note that 23MPX0134C(D2), 23MPX0135V(C2), and 23MPX0136V(C3) are samples from crust or vesicles from separate individuals. Sequences can be accessed with the following GISAID (https://www.gisaid.org) accession nos.: EPI_ISL_19287107 (D2), EPI_ISL_19287108 (C2), EPI_ISL_19287109 (C3). Scale bar indicates number of substitutions per site.
Main Article
Page created: August 28, 2024
Page updated: September 23, 2024
Page reviewed: September 23, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
