Volume 30, Number 11—November 2024
Dispatch
Co-Circulation of 2 Oropouche Virus Lineages, Amazon Basin, Colombia, 2024
Cite This Article
Citation for Media
Abstract
In early 2024, explosive outbreaks of Oropouche virus (OROV) linked to a novel lineage were documented in the Amazon Region of Brazil. We report the introduction of this lineage into Colombia and its co-circulation with another OROV lineage. Continued surveillance is needed to prevent further spread of OROV in the Americas.
Oropouche virus (OROV; Orthobunyavirus oropoucheense) is a reemerging arbovirus belonging to the family Peribunyaviridae. The large (L), medium (M), and small (S) single-stranded, negative-sense RNA segments of the OROV genome are susceptible to reassortments (1). Besides acute fever, OROV can cause meningitis and encephalitis. Although Culicoides paraensis biting midges are the main vector for OROV in urban cycles, different insect species are vectors in sylvatic cycles. Vertebrates, such as primates, small mammals, and possibly birds, are reservoir hosts (1,2).
OROV has spread rapidly in South America, causing >500,000 human infections (2,3). Most OROV cases have occurred in the Amazon in Brazil, where explosive outbreaks, sometimes affecting thousands of inhabitants, have been reported since OROV was identified in Trinidad and Tobago in 1955 (2,4,5). During 2019–2021, OROV was responsible for >10% and dengue virus (DENV) for 20% of acute febrile illness cases at 4 sites in Colombia: Cúcuta, Cali, Villavicencio, and Leticia (6). Phylogenetic analysis revealed 2 separate OROV introductions into those sites in Colombia from bordering Ecuador or Peru (6).
In early February 2024, the Pan American Health Organization and World Health Organization issued an epidemiologic alert because of the dramatic increase in OROV cases in 4 states within the Amazon Region of Brazil (7). The state of Amazonas, Brazil, alone had >1,000 quantitative reverse transcription PCR (qRT-PCR)–confirmed cases reported during 2023–January 2024 (8). After analyzing hundreds of full-length genomes isolated during the large-scale OROV outbreak in Brazil, multiple reassortment events were identified, indicating a new OROV lineage, BR-2015-2024 (9; F.C.M. Iani et al., unpub. data, https://doi.org/10.1101/2024.08.02.24311415). We report human OROV cases in Colombia caused by the novel BR-2015-2024 lineage, which is co-circulating with another previously characterized OROV lineage (6). The protocol for this study was approved by the ethics committee of the Corporación para Investigaciones Biológicas (protocol no. SC-6230-1).
In early January 2024, health authorities noticed a slight increase in acute febrile cases in Leticia municipality, which has >53,000 inhabitants, in the Amazonas department of Colombia. A total of 117 persons reporting chills (94.6%), headache (87.2%), arthralgia (65%), myalgia (41%), diarrhea (34.2%), fatigue (33.3%), and rash (6%) were treated at the emergency room of a Leticia hospital during a 5-week period. Although fever (>38.5°C) was confirmed at the doctor’s office in >46% of patients, <3% had respiratory symptoms. No hemorrhagic manifestations were observed in the febrile patients, and severe illnesses or hospitalizations were not reported. Patients were 7–92 years of age; 60 were male and 57 female. After obtaining written consent from adults and the children’s parents or legal guardians, hospital staff collected serum samples.
Frozen serum samples were shipped by air >1,300 km to One Health Colombia in Medellin, Colombia, a center established by the Global Health Institute at the University of Wisconsin, Madison (Madison, WI, USA), and the National University of Colombia-Medellin. According to center guidelines at One Health Colombia (6), we tested the serum samples for Zika virus (ZIKV), Mayaro virus (MAYV), chikungunya virus, DENV, OROV, hepatitis B and C viruses, Leptospira spp., and Plasmodium spp. (Appendix Table 1).
We detected the DENV genome in 8 samples by using the Zika, chikungunya, and dengue virus Trioplex real-time qRT-PCR (Appendix Table 1); 4 samples were positive for the DENV nonstructural protein 1, and 3 samples were positive for DENV IgM. The study population (n = 117) had a high DENV IgG prevalence of 77.7%. All samples were negative for ZIKV, CHIKV, and MAYV RNA. We detected Plasmodium spp. DNA in 7 samples by quantitative PCR, and 5 samples were positive for the P. vivax antigen. No samples were positive for hepatitis B virus surface antigen, hepatitis C virus antibody, or Leptospira spp. DNA.
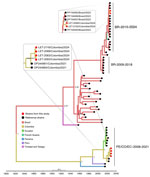
Using an OROV qRT-PCR designed at One Health Colombia (6), we detected OROV L and M segment RNA in 8 samples. We conducted next generation sequencing by using metagenomic and target enrichment approaches for those 8 serum specimens, as described previously (6). We conducted phylogenetic analysis of the OROV L, M, and S segments isolated from patient samples by using available OROV genomes and sequences of the new clade from Brazil. OROV BR-2015-2024 from Brazil (9; F.C.M. Iani et al., unpub. data) was present in 2 samples from Leticia municipality, designated as LET-2099 and LET-2102 (Figures 1, 2). Although the L and S segments of those 2 OROV samples each branched as paraphyletic clades basal to OROV PE/CO/EC-2008-2021, the M segment sequences were more closely related to OROV BR-2009-2018 sequences. Amino acid changes within lineage BR-2015-2024 indicated multiple mutations in L and M segments when compared with those in ancestral virus strains (Appendix Figure). In addition, in 6 of the 8 samples, we detected the OROV PE/CO/EC-2008-2021 lineage, which circulated in Colombia during 2019–2021 (Table) (6). The 8 OROV-positive patients did not exhibit co-infections with the other tested organisms, aside from clinical manifestations of acute undifferentiated fever (Appendix Table 2), and they had not traveled outside of Leticia municipality or its surrounding areas in the 2 weeks before symptom onset (Figure 3). We deposited 6 full-length OROV genomes analyzed in this study, including the 2 novel OROV lineages, into GenBank (accession nos. PP477303–20).
We report the circulation of the novel orthobunyavirus, OROV BR-2015-2024, in Colombia that has been documented in Brazil. The numerous mutations in OROV BR-2015-2024 RNA-dependent RNA polymerase and glycoproteins likely enhanced replication and immune evasion capabilities, increasing virus fitness and transmission. In addition, we detected co-circulation of OROV PE/CO/EC-2008-2021 lineage (6) in 6 patients. The identification of co-circulating strains of OROV exemplifies the evolving nature of orthobunyaviruses and raises concerns about future reassortment events and emergence of new lineages having more severe clinical phenotypes and enhanced vector competence. In an arbovirus hotspot, such as Leticia, it is unknown if cross-protective immunity will exist between the 2 OROV lineages. Previously, a reassortant OROV isolated from outbreaks in Iquitos, the largest city in the Amazon of Peru, provided limited cross-protection against a different OROV strain (10).
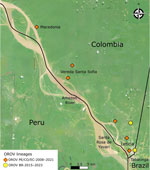
Leticia municipality, in the southernmost region in Colombia (Figure 3), remains isolated from the rest of Colombia’s road network, and travelers typically reach Leticia by aircraft. Leticia city, the capital of Amazonas department of Colombia, is in Leticia municipality, where the borders of Colombia, Brazil, and Peru converge, forming an area known as the Three Borders. Tabatinga city, in the state of Amazonas in Brazil, is located across from Leticia city, forming a unique suburban area near Santa Rosa de Yavarí, Peru, which is on an island in the Amazon River (11). The new OROV lineage is likely spreading rapidly in those borderlands because of the considerable human mobility related to local businesses. Furthermore, we believe that the novel OROV lineage in Leticia came from either a nearby settlement or air travelers from the Amazon in Brazil, where persistent outbreaks are occurring.
One Health Colombia has previously reported outbreaks of ZIKV (12), DENV (13), OROV (6), and, in 2024, MAYV (14) in Colombia, which underscores the dynamic landscape of reemerging arboviruses in the region. The sustained transmission of established strains and the unpredictable reassortment events of OROVs causing outbreaks is yet another demonstration of the public health challenges associated with prevention and control of arbovirus reemergence in South America. Continued surveillance and molecular characterization of OROV and other arboviruses are needed to prevent the spread of arboviral diseases in the Americas.
Mr. Usuga is a research scientist and bioinformatician at the One Health Colombia center at the National University of Colombia in Medellin. His research interests focus on data modeling, genomics, and molecular epidemiology of emerging and reemerging viral diseases.
Acknowledgments
We thank the clinical and laboratory staff at the Hospital San Rafael de Leticia and Fundación Clínica Leticia for their participation in our study; the staff of One Health Colombia, particularly the laboratory personnel, for their support of this work; Fundación Fraternidad Medellin and Fundación Sofia Perez de Soto for their unwavering support for One Health PhD awards; and Celeny Ortiz Restrepo for generating the map of Leticia municipality in the Amazon Basin, Colombia.
This research was supported by the Global Health Institute of the University of Wisconsin-Madison, the National University of Colombia-Medellin, and the virus discovery research program of the Abbott Pandemic Defense Coalition.
References
- Wesselmann KM, Postigo-Hidalgo I, Pezzi L, de Oliveira-Filho EF, Fischer C, de Lamballerie X, et al. Emergence of Oropouche fever in Latin America: a narrative review. Lancet Infect Dis. 2024;24:e439–52. DOIPubMedGoogle Scholar
- Pinheiro FP, Travassos da Rosa AP, Travassos da Rosa JF, Ishak R, Freitas RB, Gomes ML, et al. Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am J Trop Med Hyg. 1981;30:149–60. DOIPubMedGoogle Scholar
- Forshey BM, Guevara C, Laguna-Torres VA, Cespedes M, Vargas J, Gianella A, et al.; NMRCD Febrile Surveillance Working Group. Arboviral etiologies of acute febrile illnesses in Western South America, 2000-2007. PLoS Negl Trop Dis. 2010;4:
e787 . DOIPubMedGoogle Scholar - Anderson CR, Spence L, Downs WG, Aitken TH. Oropouche virus: a new human disease agent from Trinidad, West Indies. Am J Trop Med Hyg. 1961;10:574–8. DOIPubMedGoogle Scholar
- Azevedo RSS, Nunes MR, Chiang JO, Bensabath G, Vasconcelos HB, Pinto AY, et al. Reemergence of Oropouche fever, northern Brazil. Emerg Infect Dis. 2007;13:912–5. DOIPubMedGoogle Scholar
- Ciuoderis KA, Berg MG, Perez LJ, Hadji A, Perez-Restrepo LS, Aristizabal LC, et al. Oropouche virus as an emerging cause of acute febrile illness in Colombia. Emerg Microbes Infect. 2022;11:2645–57. DOIPubMedGoogle Scholar
- Pan American Health Organization, World Health Organization. Epidemiological alert—Oropouche in the Region of the Americas—2 February 2024 [cited 2024 Mar 10]. https://www.paho.org/en/documents/epidemiological-alert-oropouche-region-americas-2-february-2024
- Pan American Health Organization, World Health Organization. Public health risk assessment related to Oropouche virus (OROV) in the Region of the Americas—9 February 2024 [cited 2024 Mar 10]. https://www.paho.org/en/documents/public-health-risk-assessment-related-oropouche-virus-orov-region-americas-9-february
- Naveca FG, de Almeida TAP, Souza V, Nascimento V, Silva D, Nascimento F, et al. Human outbreaks of a novel reassortant Oropouche virus in the Brazilian Amazon region. Nat Med. 2024; Epub ahead of print. DOIPubMedGoogle Scholar
- Aguilar PV, Barrett AD, Saeed MF, Watts DM, Russell K, Guevara C, et al. Iquitos virus: a novel reassortant Orthobunyavirus associated with human illness in Peru. PLoS Negl Trop Dis. 2011;5:
e1315 . DOIPubMedGoogle Scholar - Farnsworth-Alvear A, Palacios M, López AMG, editors. The Colombia reader: history, culture, politics. Durham (NC): Duke University Press; 2016.
- Camacho E, Paternina-Gomez M, Blanco PJ, Osorio JE, Aliota MT. Detection of autochthonous Zika virus transmission in Sincelejo, Colombia. Emerg Infect Dis. 2016;22:927–9. DOIPubMedGoogle Scholar
- Ciuoderis KA, Usuga J, Moreno I, Perez-Restrepo LS, Flórez DY, Cardona A, et al. Characterization of dengue virus serotype 2 cosmopolitan genotype circulating in Colombia. Am J Trop Med Hyg. 2023;109:1298–302. DOIPubMedGoogle Scholar
- Perez-Restrepo LS, Ciuoderis K, Usuga J, Moreno I, Vargas V, Arévalo-Arbelaez AJ, et al. Mayaro virus as the cause of acute febrile illness in the Colombian Amazon Basin. Front Microbiol. 2024;15:
1419637 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleOriginal Publication Date: October 02, 2024
1These first authors contributed equally to this article.
Table of Contents – Volume 30, Number 11—November 2024
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Jorge E. Osorio, Global Health Institute, University of Wisconsin, 1050 MSC, 1300 University Ave, Madison, WI 53706, USA
Top