Volume 30, Number 11—November 2024
Dispatch
Co-Circulation of 2 Oropouche Virus Lineages, Amazon Basin, Colombia, 2024
Figure 2
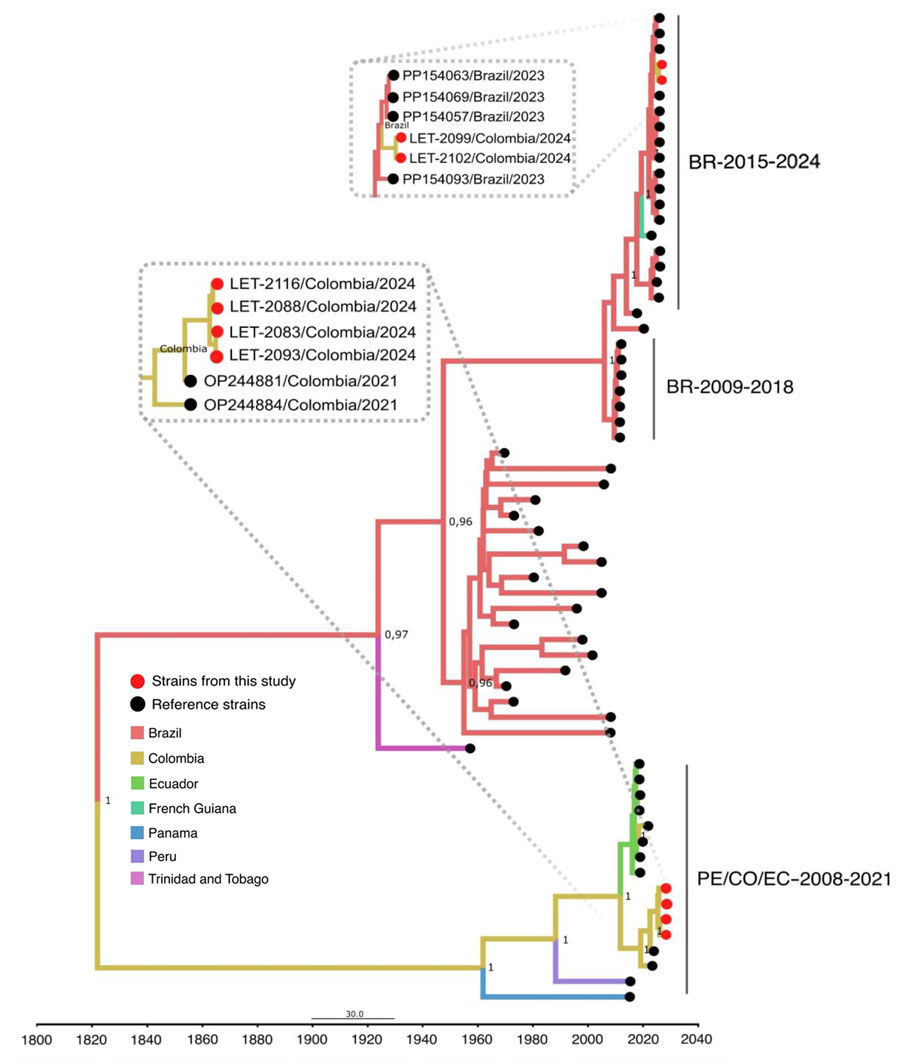
Figure 2. Time-scaled Bayesian phylogeographic analysis of medium segments of co-circulating Oropouche virus lineages, Amazon Basin, Colombia, 2024. Bayesian phylogenetic tree of medium gene segments were estimated by using the Bayesian Markov chain Monte Carlo method (>100 million generations) in Beast (https://beast.community) and ModelFinder in IQ-TREE (http://www.iqtree.org) (ultrafast bootstrapping and 1,000 replicates). Red solid circles indicate viruses from this study that begin with LET for Leticia, Colombia. Phylogeny branches are colored according to their descendant place of origin. Best-fit model was selected according to Bayesian information criteria, and uncorrelated relaxed molecular clock model was used. Bayesian posterior values (>0.8) are annotated at specific nodes of the trees. Sequences from this study were compared with reference sequences from other studies. Main clusters are indicated by using the following reference labels: BR-2015-2024 cluster represents the recent outbreak of the new OROV lineage in Brazil during 2015–2024; PE/CO/EC-2008-2021 cluster represents sequences from Colombia, Peru, and Ecuador during 2008–2021; and BR-2009-2018 cluster represents sequences from Brazil during 2009–2018. Scale bar indicates nucleotide substitutions per site.
1These first authors contributed equally to this article.