Volume 30, Supplement—March 2024
SUPPLEMENT ISSUE
Outbreaks and Investigations
Outbreak of Invasive Serratia marcescens among Persons Incarcerated in a State Prison, California, USA, March 2020–December 2022
Figure 3
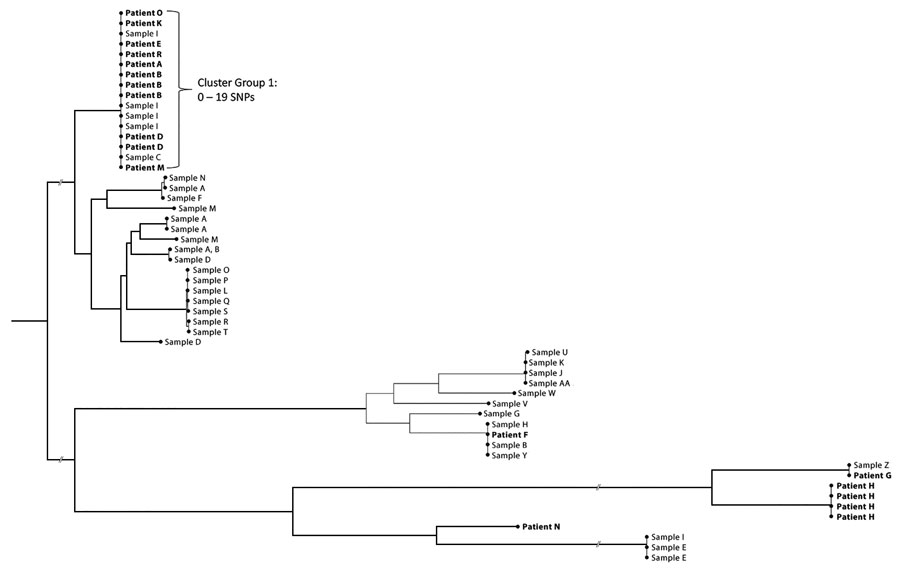
Figure 3. Phylogenetic tree representing patients hospitalized with invasive Serratia marcescens infections and whole-genome sequencing for environmental and clinical isolates at prison A, California, USA, January 2020–March 2023. The predominant outbreak cluster included patients A, B, D, E, K, M, O, and R and environmental samples C (needle/syringe) and sample I (nasal spray bottle) from patient D. These sequences had 0–19 single-nucleotide polymorphism (SNP) differences. Patient F, sample B (coffee cup) found in patient D’s cell, sample H (hand rinsate) from the cellmate of patient D, and sample Y (doorway swab) from the cell occupied at different times by both patient A and AC are grouped together within a 11–17 SNP range.
Page created: October 12, 2023
Page updated: March 31, 2024
Page reviewed: March 31, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.