Molecular Characterization of Noroviruses Causing Acute Gastroenteritis Outbreaks among US Military Recruits, 2013–2023
Melissa S. Anderson, Chanel A. Mosby-Tourtellot, Regina Z. Cer, Daniel J. Crouch, Ryan S. Underwood, Cailin S. La Claire, Roger W. Pan, Evelyn W. Mahugu, Hunter J. Smith, Kimberly A. Bishop-Lilly, Mathew D. Esona, Francisco Mimica Porras

, and Michelle Hartman-Lane
Author affiliation: Naval Health Research Center, Operational Infectious Diseases Directorate, San Diego, California, USA (M.S. Anderson, D.J. Crouch, R.S. Underwood, C.A. La Claire, R.W. Pan, M.D. Esona, F.M. Porras, M. Hartman-Lane); General Dynamics Information Technology, San Diego (M.S. Anderson, R.S. Underwood, C.A. La Claire, R.W. Pan); Defense Threat Reduction Agency, Fort Belvoir, Virginia, USA (C.A. Mosby-Tourtellot); Naval Medical Research Command, Biological Defense Research Directorate, Fort Detrick, Maryland, USA (C.A. Mosby-Tourtellot, R.Z. Cer, K.A. Bishop-Lilly); General Dynamics, Silver Spring, Maryland, USA (E.W. Mahugu); Global Emerging Infections Surveillance Branch, Armed Forces Health Surveillance Division, Silver Spring (E.W. Mahugu, H.J. Smith); Uniformed Services University, Bethesda, Maryland, USA (H.J. Smith).
Main Article
Figure 2
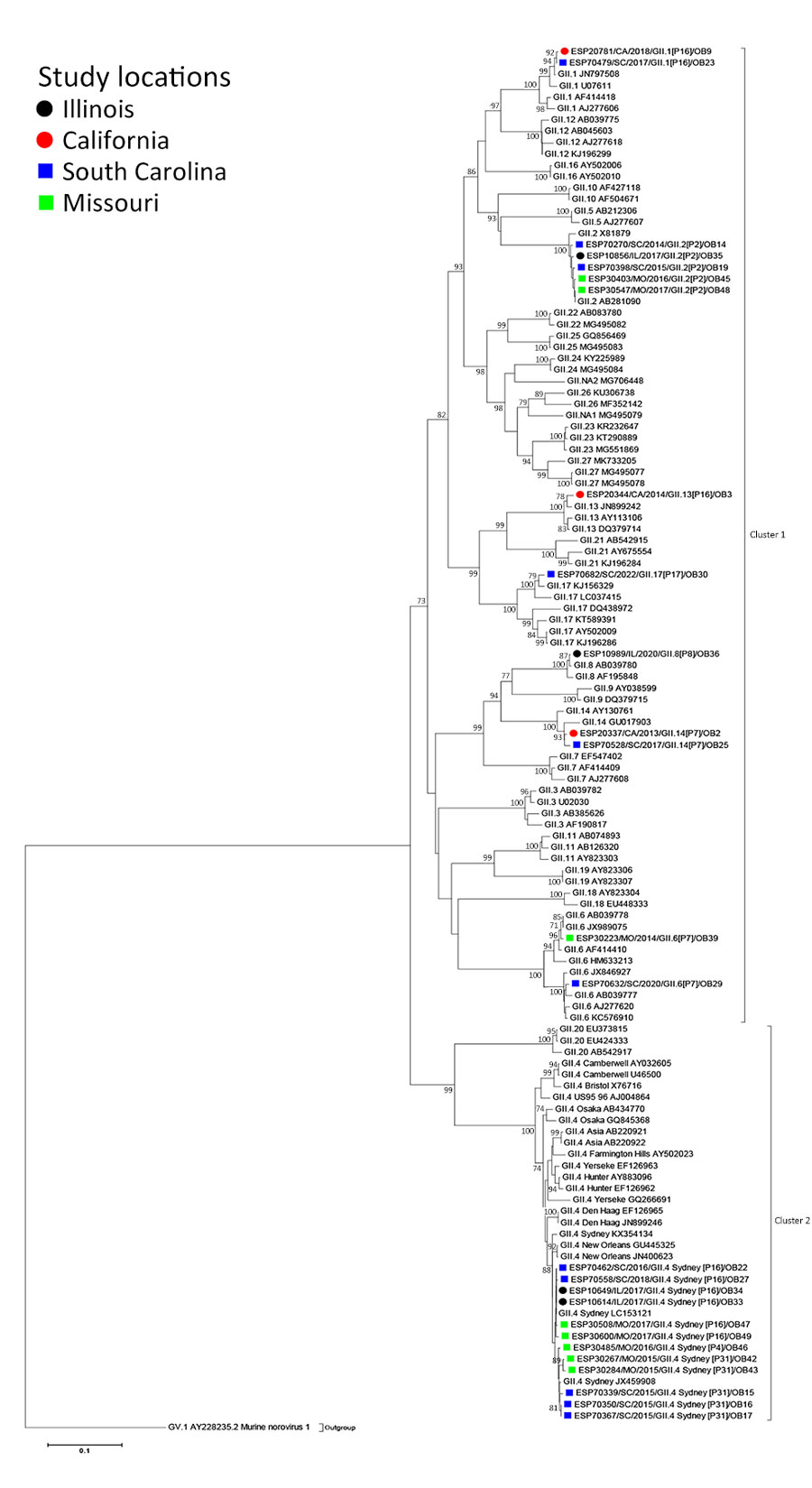
Figure 2. Maximum-likelihood phylogram of GI ORF2 (VP1) in a study of molecular characterization of noroviruses causing acute gastroenteritis outbreaks among US military recruits at 4 basic training facilities, 2013–2023. Phylogram reveals genetic relatedness of GI ORF2 (VP1) deduced amino acid sequences (+550 bp) for 13 of the 39 norovirus outbreak strains. Outbreak study strains are labeled from left to right as follows: sample identification/location sample was collected/year sample was collected/genogroup and P-type/outbreak number. The 4 facilities, identified by color, were Marine Corps centers in California and South Carolina, a Navy center in Illinois, and an Army center in Missouri. Bootstrap values >70% are indicated at branch nodes where applicable. Scale bar indicates nucleotide substitutions per site. ORF, open reading frame; VP, virus capsid protein.
Main Article
Page created: September 18, 2024
Page updated: November 11, 2024
Page reviewed: November 11, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
