Volume 30, Number 2—February 2024
Research Letter
Fatal West Nile Virus Infection in Horse Returning to United Kingdom from Spain, 2022
Figure 1
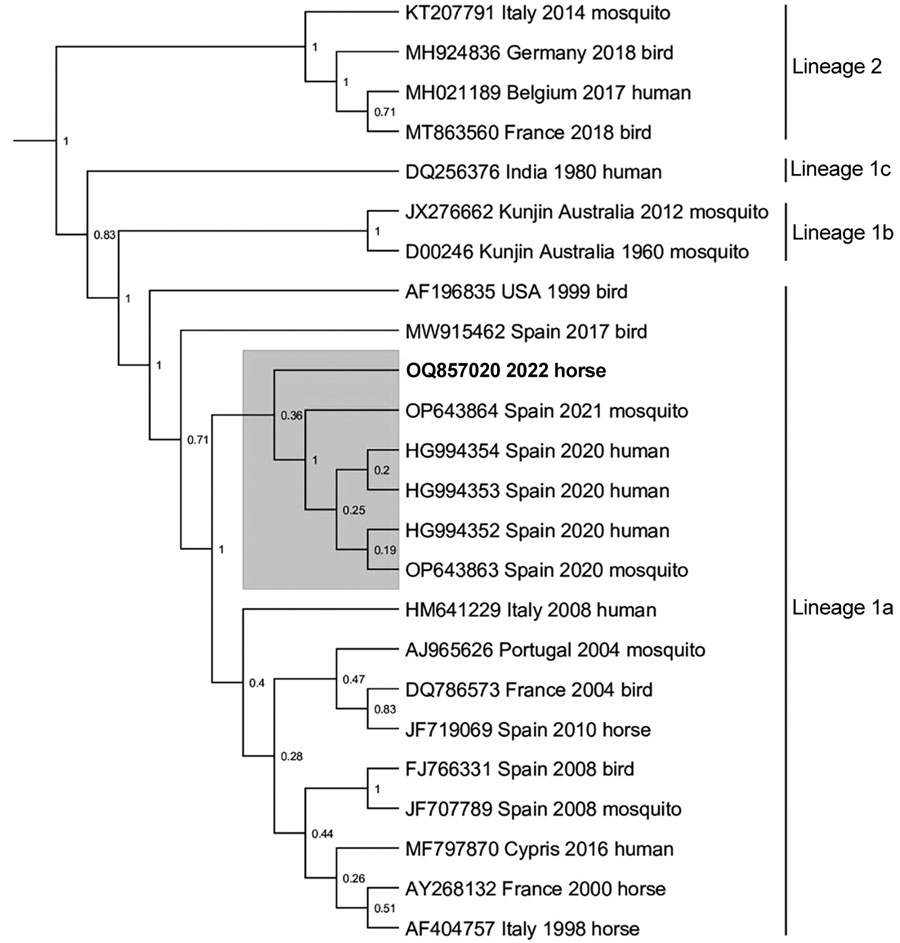
Figure 1. Next-generation sequencing data associated with fatal West Nile virus (WNV) infection in a horse returning to the United Kingdom from Spain, 2022. Bayesian phylogenetic tree analysis of a 624-bp sequence located in the nonstructural 5 gene showed that the strain from the horse (boldface; GenBank accession no. OQ857020) clusters with recent sequences from Andalusia, Spain (gray shading). Next-generation sequencing was conducted on an Illumina MiSeq sequencer (https://www.illumina.com). The sequence was aligned with 23 WNV lineage 1 and 2 reference sequences obtained from GenBank in MEGA version 11.0.13 (https://www.megasoftware.net), and a Bayesian phylogenetic analysis was undertaken in BEAST version 1.10.4 (https://beast.community) using a general time reversible plus invariant sites plus gamma nucleotide substitution model and 10,000,000 Markov chain Monte Carlo generations. Node labels represent posterior probabilities. Accession number, country, year of detection and host species are included for each sequence.