Volume 30, Number 2—February 2024
Research Letter
Lymphocytic Choriomeningitis Virus Lineage V in Wood Mice, Germany
Figure
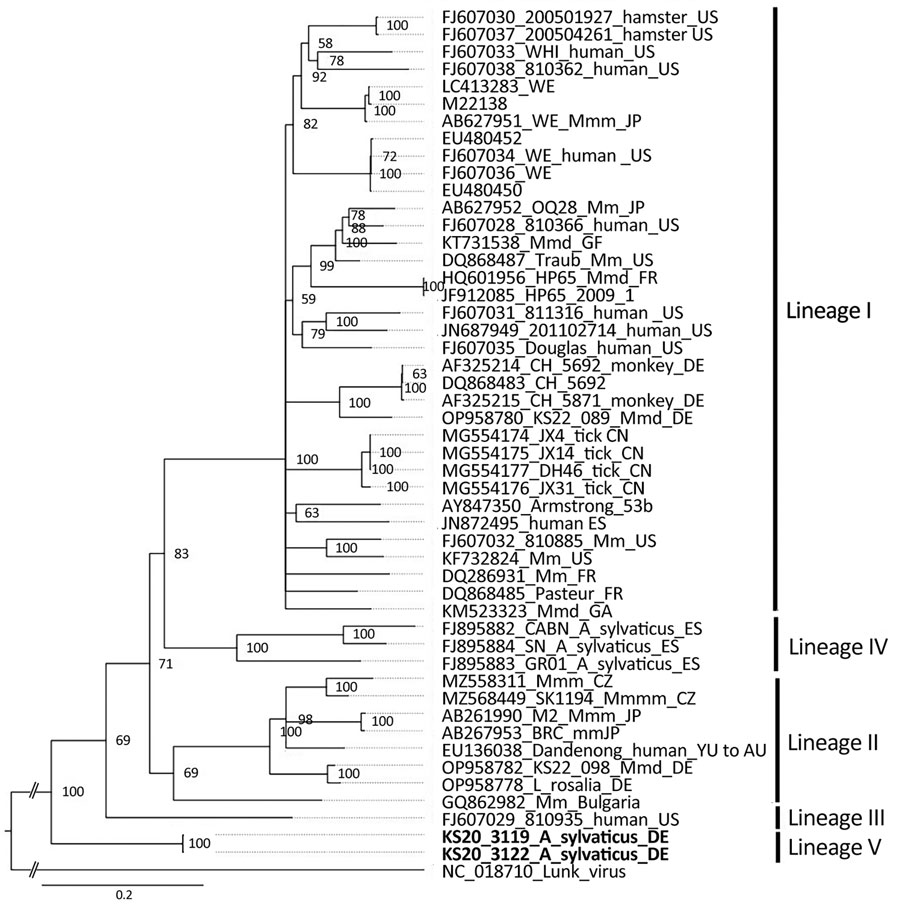
Figure. Phylogenetic analysis of the nucleocapsid protein encoding region of lymphocytic choriomeningitis virus lineage V identified in wood mice, Germany (boldface), and reference sequences. Bayesian inference method was used to analyze the 1,674-nt open reading frame corresponding to codons 1–558 without the stop codon. GenBank accession number, strain name, host species, and country of origin (if known) are shown. Roman numerals I–IV represent the different virus lineages as defined previously (10). Lunk virus from Mus minutoides mice was used as an outgroup. WE and Armstrong are laboratory strains of lymphocytic choriomeningitis virus. Scale bar indicates nucleotide substitutions per site. Asyl, Apodemus sylvaticus; AU, Australia; BG, Bulgaria; CN, China; CZ, Czech Republic; DE, Germany; ES, Spain; FR, France; GA, Gabon; GF, French Guiana; JP, Japan; Mm, Mus musculus; Mmm, M. musculus musculus; Mmd, M. musculus domesticus; SK, Slovakia; US, United States; YU, former Yugoslavia.
References
- Meyer BJ, de la Torre JC, Southern PJ. Arenaviruses: genomic RNAs, transcription, and replication. Curr Top Microbiol Immunol. 2002;262:139–57. DOIPubMedGoogle Scholar
- Ackermann R, Stille W, Blumenthal W, Helm EB, Keller K, Baldus O. [Syrian hamsters as vectors of lymphocytic choriomeningitis] [in German]. Dtsch Med Wochenschr. 1972;97:1725–31. DOIPubMedGoogle Scholar
- Goldwater PN. A mouse zoonotic virus (LCMV): A possible candidate in the causation of SIDS. Med Hypotheses. 2021;158:
110735 . DOIPubMedGoogle Scholar - Asper M, Hofmann P, Osmann C, Funk J, Metzger C, Bruns M, et al. First outbreak of callitrichid hepatitis in Germany: genetic characterization of the causative lymphocytic choriomeningitis virus strains. Virology. 2001;284:203–13. DOIPubMedGoogle Scholar
- Ledesma J, Fedele CG, Carro F, Lledó L, Sánchez-Seco MP, Tenorio A, et al. Independent lineage of lymphocytic choriomeningitis virus in wood mice (Apodemus sylvaticus), Spain. Emerg Infect Dis. 2009;15:1677–80. DOIPubMedGoogle Scholar
- Schmidt S, Essbauer SS, Mayer-Scholl A, Poppert S, Schmidt-Chanasit J, Klempa B, et al. Multiple infections of rodents with zoonotic pathogens in Austria. Vector Borne Zoonotic Dis. 2014;14:467–75. DOIPubMedGoogle Scholar
- Kallio-Kokko H, Laakkonen J, Rizzoli A, Tagliapietra V, Cattadori I, Perkins SE, et al. Hantavirus and arenavirus antibody prevalence in rodents and humans in Trentino, Northern Italy. Epidemiol Infect. 2006;134:830–6. DOIPubMedGoogle Scholar
- Mehl C, Wylezich C, Geiger C, Schauerte N, Mätz-Rensing K, Nesseler A, et al. Reemergence of lymphocytic choriomeningitis mammarenavirus, Germany. Emerg Infect Dis. 2023;29:631–4. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, et al. RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Albariño CG, Palacios G, Khristova ML, Erickson BR, Carroll SA, Comer JA, et al. High diversity and ancient common ancestry of lymphocytic choriomeningitis virus. Emerg Infect Dis. 2010;16:1093–100. DOIPubMedGoogle Scholar
1Current affiliation: Friedrich-Loeffler-Institut, Greifswald-Insel Riems, Germany and Justus-Liebig-Universität Gießen, Gießen, Germany.