High Prevalence of Echinostoma mekongi Infection in Schoolchildren and Adults, Kandal Province, Cambodia
Bong-Kwang Jung
1, Taehee Chang
1, Seungwan Ryoo, Sooji Hong, Jeonggyu Lee, Sung-Jong Hong, Woon-Mok Sohn, Virak Khieu, Rekol Huy, and Jong-Yil Chai

Author affiliations: MediCheck Research Institute, Korea Association of Health Promotion, Seoul, South Korea (B.-K. Jung, S. Ryoo, S. Hong, J. Lee); Seoul National University Graduate School of Public Health, Seoul (T. Chang); Seoul National University College of Medicine, Seoul (J.-Y. Chai); Convergence Research Center for Insect Vectors, Inchon National University, Incheon, South Korea (S.-J. Hong); Gyeongsang National University College of Medicine, Jinju, South Korea (W.-M. Sohn); National Center for Parasitology, Entomology and Malaria Control, Ministry of Health, Phnom Penh, Cambodia (V. Khieu, R. Huy)
Main Article
Figure 2
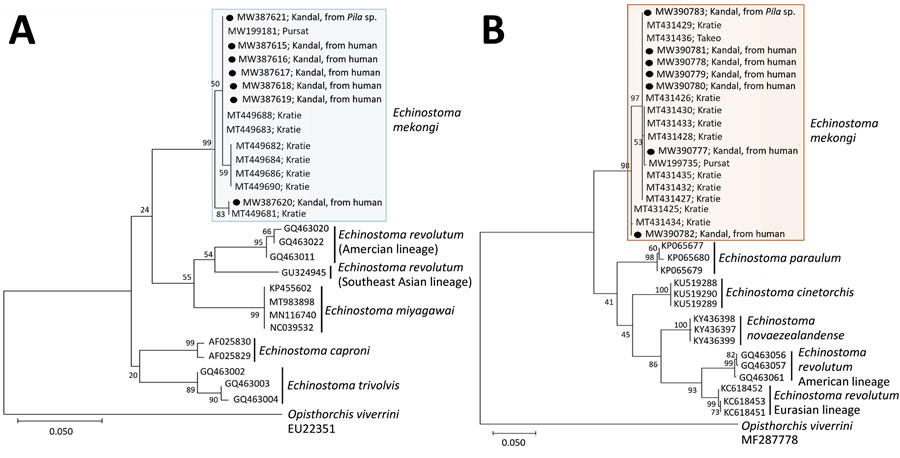
Figure 2. Phylogenetic trees of cox1 (A) and nd1 (B) genes of Echinostoma mekongi adults (n = 6) extracted from volunteers and metacercaria (n = 1) extracted from Pila sp. snails for study of E. mekongi infection in schoolchildren and adults, Kandal Province, Cambodia. Sequences from this study (shades boxes) are shown in comparison with other 37-collar-spined Echinostoma spp. (outgroup; Opisthorchis viverrini). The trees were constructed using the maximum-likelihood method, employing the Tamura-Nei model of nucleotide substitution with 1,000 bootstrap replications and viewed in MEGA X (https://www.megasoftware.net). GenBank accession numbers are given for all sequences. Scale bars indicate substitutions per site.
Main Article
Page created: January 31, 2024
Page updated: February 22, 2024
Page reviewed: February 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
