Volume 30, Number 4—April 2024
Research
A One Health Perspective on Salmonella enterica Serovar Infantis, an Emerging Human Multidrug-Resistant Pathogen
Figure 2
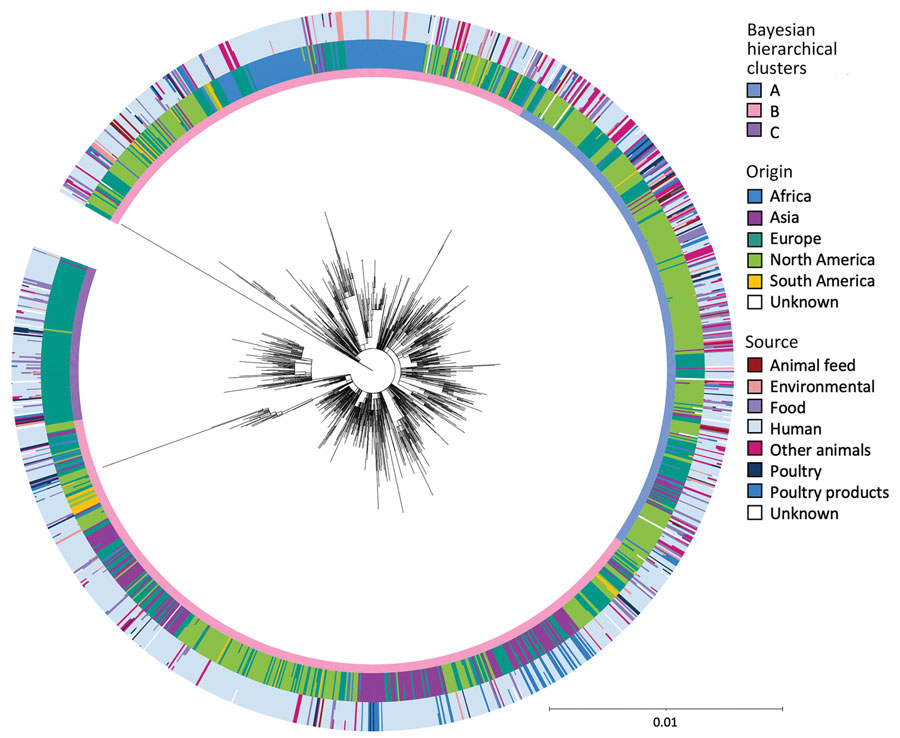
Figure 2. Core single-nucleotide polymorphism maximum-likelihood phylogeny of 1,288 representatives of 5,283 isolates in One Health–focused analysis of emerging multidrug-resistant pathogen Salmonella enterica serovar Infantis. The inner ring around the phylogeny is annotated with the Bayesian hierarchical clusters found by fastbaps. Cluster A consists of 348 representatives of 1,624 isolates; cluster B consists of 831 representatives of 3,283 isolates; and cluster C consists of 109 representatives of 376 isolates. The outer rings show the percentage of isolates in each 25-SNP cluster that were from each continent and source. Isolate origin was identified for Africa (n = 316), Asia (n = 312), Europe (n = 1,641), North America (n = 2,861), and South America (n = 128); the origin of the remaining isolates was unknown (n = 25). Sources were animal feed (n = 74), environmental (n = 268), food (n = 390), human (n = 3,149), other animals (n = 321), poultry (n = 300), poultry products (n = 684), and unknown (n = 97).
1Current affiliation: University of Edinburgh, Edinburgh, Scotland, UK.
2Current affiliation: Utrecht University, Utrecht, the Netherlands.
3Current affiliation: UK Health Security Agency, London, UK.
4Current affiliation: European Bioinformatics Institute, Cambridge, UK.