Volume 30, Number 7—July 2024
Dispatch
Multicountry Spread of Influenza A(H1N1)pdm09 Viruses with Reduced Oseltamivir Inhibition, May 2023–February 2024
Figure 1
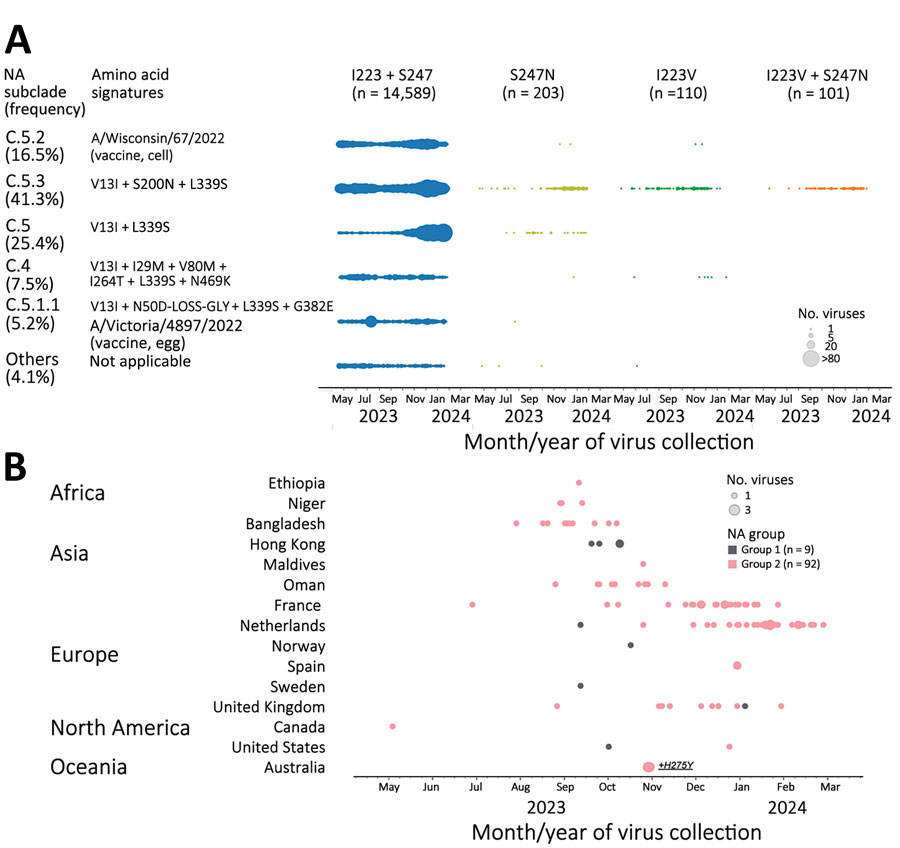
Figure 1. Detection of influenza A(H1N1)pdm09 viruses with dual NA-I223V + S247N substitutions through NA inhibitors susceptibility surveillance conducted by the Centers for Disease Control and Prevention and analysis of available sequences (GISAID EpiFlu, https://www.gisaid.org, accessed March 11, 2024), May 2023–February 2024. A total of 15,003 NA sequences of pH1N1 viruses (duplicate sequences excluded: 2,039 from Centers for Disease Control and Prevention and the remaining 12,964 from GISAID EpiFlu) were analyzed to screen for amino acid substitutions at residues 223 and 247. A) Introduction of single substitution (I223V or S247N) or dual substitutions (I223V + S247N) across NA subclades of pH1N1 viruses circulating during May 2023–February 2024. Amino acid signatures of NA subclades are shown in comparison to A/Wisconsin/67/2022, the Northern Hemisphere 2023–2024 vaccine cell prototype virus for the pH1N1 component. Vaccine viruses, A/Wisconsin/67/2022 and A/Victoria/4897/2022 (Northern Hemisphere 2023–2024 vaccine egg prototype virus), represented NA subclades C.5.2 and C.5.1.1, respectively. C.5.3 was the subclade most abundantly sequenced (41.3% frequency), followed by other minor subclades: C.5 (25.4%), C.5.2 (16.5%), C.4 (7.5%), C.5.1.1 (5.2%), and others (4.1%). Most viruses with single S247N substitution belonged to dominant NA subclade C.5.3 and minor subclade C.5. Conversely, most viruses with single I223V substitution and all viruses with dual I223V + S247N substitutions belonged to NA subclade C.5.3. B) Spatiotemporal distribution of dual mutant viruses. Dual mutants were divided into 2 groups based on their NA sequence difference. Group 1 shared additional substitution R257K not found in group 2. The small group 1 had 9 dual mutants, and the large group 2 had 92 dual mutants. The first dual mutant belonging to group 2 was collected in Canada at the end of May 2023, and most dual mutants were collected between September 2023 and February 2024. Two dual mutant viruses collected in Australia also contained NA-H275Y. NA, neuraminidase.
1These authors contributed equally to this article.