Volume 30, Number 9—September 2024
Dispatch
Zoonotic Mansonella ozzardi in Raccoons, Costa Rica, 2019–2022
Figure
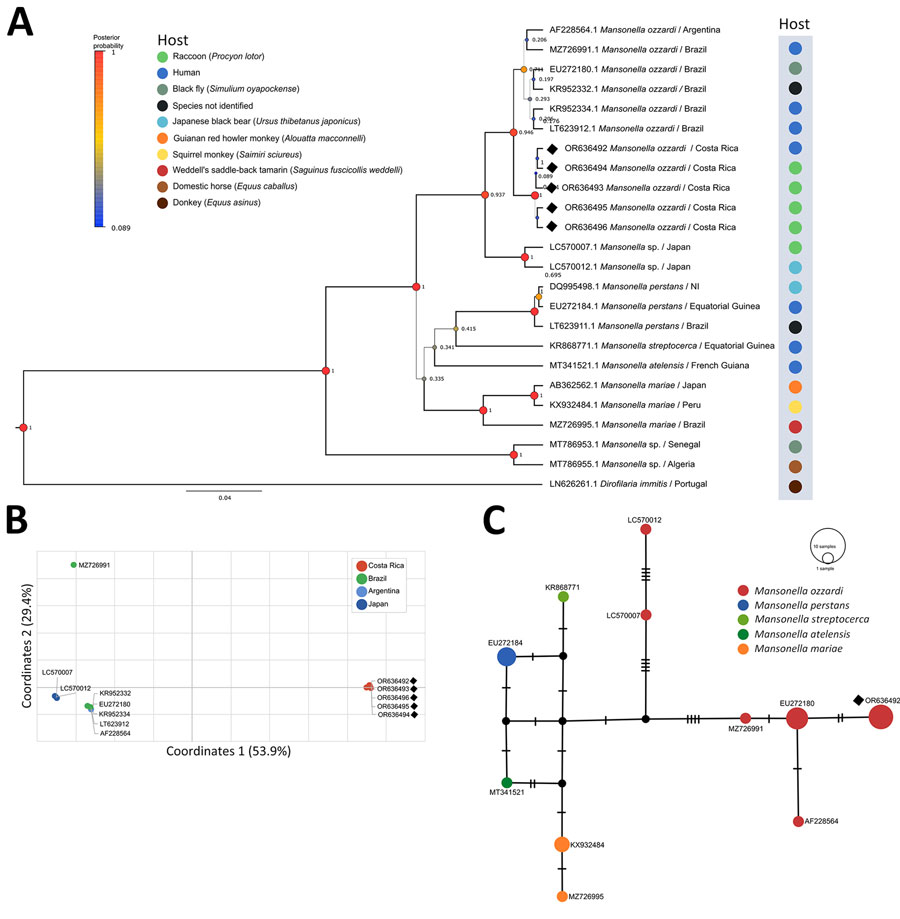
Figure. Phylogenetic and haplotypic analysis of Mansonella ozzardi ITS1 sequences obtained from racoons (Procyon lotor) in Costa Rica, 2019–2022 (black diamonds). A) Bayesian inference phylogenetic tree of Mansonella spp. based on the Hasegawa-Kishino-Yano with gamma distribution model. Line thickness and node size are proportional to posterior probability values. B) Principal component analysis of Nei’s genetic distance of M. ozzardi sequences from different geographic locations. C) Templeton Crandall Sing haplotype network of Mansonella spp. sequences. Black circles indicate hypothetical haplotypes; hatchmarks indicate mutational steps between haplotypes. GenBank accession numbers are shown for all sequences.
Page created: July 18, 2024
Page updated: August 21, 2024
Page reviewed: August 21, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.