Volume 31, Number 1—January 2025
Research
Neisseria meningitidis Serogroup Y Sequence Type 1466 and Urogenital Infections
Figure 1
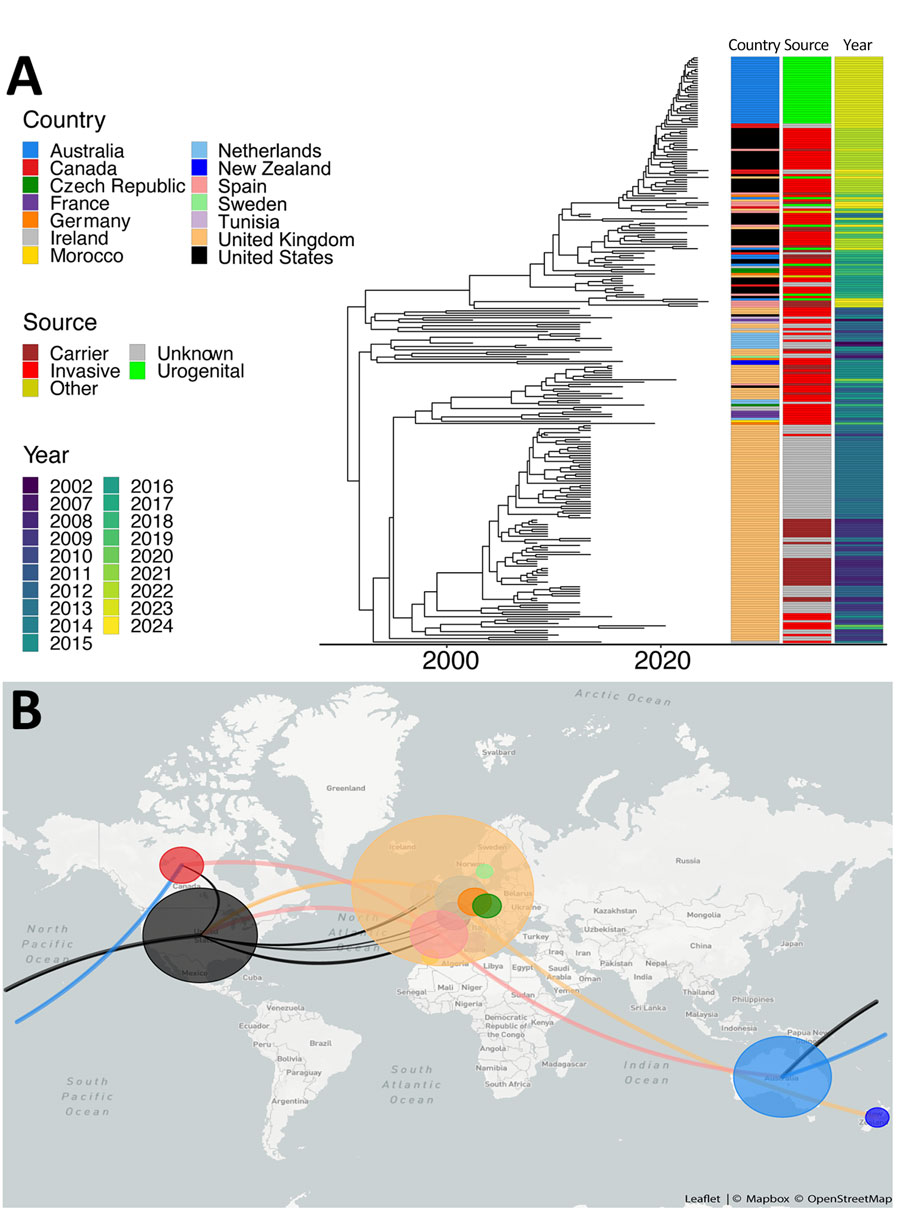
Figure 1. Relatedness of Neisseria meningitidis serogroup Y ST1466 isolates from Australia and the United States compared with isolates from other countries. A) Timed maximum-likelihood phylogeny of included isolates of serogroup Y ST1466. Associated metadata are shown to the right of the tree. B) Genomic epidemiology of ST1466 showing transmission lines generated using Nextstrain (24). ST, sequence type.
References
- Vázquez JA, de la Fuente L, Berron S, O’Rourke M, Smith NH, Zhou J, et al. Ecological separation and genetic isolation of Neisseria gonorrhoeae and Neisseria meningitidis. Curr Biol. 1993;3:567–72. DOIPubMedGoogle Scholar
- Caugant DA, Brynildsrud OB. Neisseria meningitidis: using genomics to understand diversity, evolution and pathogenesis. Nat Rev Microbiol. 2020;18:84–96. DOIPubMedGoogle Scholar
- Quillin SJ, Seifert HS. Neisseria gonorrhoeae host adaptation and pathogenesis. Nat Rev Microbiol. 2018;16:226–40. DOIPubMedGoogle Scholar
- Ladhani SN, Lucidarme J, Parikh SR, Campbell H, Borrow R, Ramsay ME. Meningococcal disease and sexual transmission: urogenital and anorectal infections and invasive disease due to Neisseria meningitidis. Lancet. 2020;395:1865–77. DOIPubMedGoogle Scholar
- Carpenter CM, Charles R. Isolation of Meningococcus from the genitourinary tract of seven patients. Am J Public Health Nations Health. 1942;32:640–3. DOIPubMedGoogle Scholar
- Tzeng YL, Bazan JA, Turner AN, Wang X, Retchless AC, Read TD, et al. Emergence of a new Neisseria meningitidis clonal complex 11 lineage 11.2 clade as an effective urogenital pathogen. Proc Natl Acad Sci U S A. 2017;114:4237–42. DOIPubMedGoogle Scholar
- Brooks A, Lucidarme J, Campbell H, Campbell L, Fifer H, Gray S, et al. Detection of the United States Neisseria meningitidis urethritis clade in the United Kingdom, August and December 2019 - emergence of multiple antibiotic resistance calls for vigilance. Euro Surveill. 2020;25:
2000375 . DOIPubMedGoogle Scholar - Takahashi H, Morita M, Yasuda M, Ohama Y, Kobori Y, Kojima M, et al. Detection of novel US Neisseria meningitidis urethritis clade subtypes in Japan. Emerg Infect Dis. 2023;29:2210–7. DOIPubMedGoogle Scholar
- Rodriguez EI, Tzeng YL, Stephens DS. Continuing genomic evolution of the Neisseria meningitidis cc11.2 urethritis clade, NmUC: a narrative review. Microb Genom. 2023;9:
001113 . DOIPubMedGoogle Scholar - Oliver SE, Retchless AC, Blain AE, McNamara LA, Ahrabifard S, Farley M, et al. Risk factors for invasive meningococcal disease belonging to a novel urethritis clade of Neisseria meningitidis—United States, 2013–2017. Open Forum Infect Dis. 2022;9:
ofac035 . DOIPubMedGoogle Scholar - McNamara LA, Potts CC, Blain A, Topaz N, Apostol M, Alden NB, et al. Invasive meningococcal disease due to nongroupable Neisseria meningitidis—active bacterial core surveillance sites, 2011–2016. Open Forum Infect Dis. 2019;6:
ofz190 . DOIPubMedGoogle Scholar - Retchless AC, Chen A, Chang HY, Blain AE, McNamara LA, Mustapha MM, et al. Using Neisseria meningitidis genomic diversity to inform outbreak strain identification. PLoS Pathog. 2021;17:
e1009586 . DOIPubMedGoogle Scholar - Lahra MM, Latham NH, Templeton DJ, Read P, Carmody C, Ryder N, et al. Investigation and response to an outbreak of Neisseria meningitidis serogroup Y ST-1466 urogenital infections, Australia. Commun Dis Intell (2018). 2024;48:48. DOIPubMedGoogle Scholar
- Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–90. DOIPubMedGoogle Scholar
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Rubis AB, Howie RL, Marasini D, Sharma S, Marjuki H, McNamara LA. Notes from the field: increase in meningococcal disease among persons with HIV—United States, 2022. MMWR Morb Mortal Wkly Rep. 2023;72:663–4. DOIPubMedGoogle Scholar
- Marjuki H, Topaz N, Rodriguez-Rivera LD, Ramos E, Potts CC, Chen A, et al. Whole-genome sequencing for characterization of capsule locus and prediction of serogroup of invasive meningococcal isolates. J Clin Microbiol. 2019;57:e01609–18. DOIPubMedGoogle Scholar
- Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:
e1005595 . DOIPubMedGoogle Scholar - Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Didelot X, Croucher NJ, Bentley SD, Harris SR, Wilson DJ. Bayesian inference of ancestral dates on bacterial phylogenetic trees. Nucleic Acids Res. 2018;46:
e134 . DOIPubMedGoogle Scholar - Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3. DOIPubMedGoogle Scholar
- Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. DOIPubMedGoogle Scholar
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–9. DOIPubMedGoogle Scholar
- Brynildsrud O, Bohlin J, Scheffer L, Eldholm V. Rapid scoring of genes in microbial pan-genome-wide association studies with Scoary. Genome Biol. 2016;17:238. DOIPubMedGoogle Scholar
- Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90. DOIPubMedGoogle Scholar
- Leong LEX, Coldbeck-Shackley RC, McMillan M, Bratcher HB, Turra M, Lawrence A, et al. The genomic epidemiology of Neisseria meningitidis carriage from a randomised controlled trial of 4CMenB vaccination in an asymptomatic adolescent population. Lancet Reg Health West Pac. 2023;43:
100966 . DOIPubMedGoogle Scholar - Workowski KA, Bachmann LH, Chan PA, Johnston CM, Muzny CA, Park I, et al. Sexually transmitted infections treatment guidelines, 2021. MMWR Recomm Rep. 2021;70:1–187. DOIPubMedGoogle Scholar
Page created: December 04, 2024
Page updated: December 22, 2024
Page reviewed: December 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.