Volume 31, Number 1—January 2025
Dispatch
Influenza A(H5N1) Virus Clade 2.3.2.1a in Traveler Returning to Australia from India, 2024
Figure 1
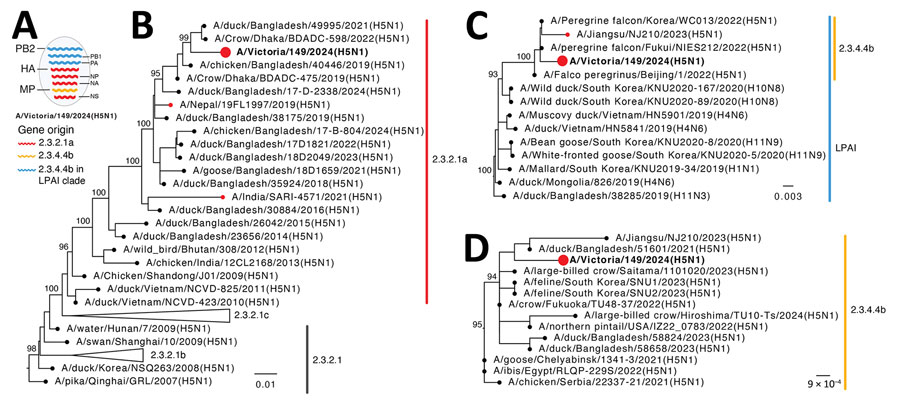
Figure 1. Evolutionary origins of isolate A/Victoria/149/2024(H5N1) (red circles and bold text) in study of influenza A(H5N1) virus clade 2.3.2.1a in traveler returning to Australia from India, 2024. A) Reassortant origins of A/Victoria/149/2024 based on analysis of each segment; detailed phylogenies for all segments are provided in Appendix Figure 1). B–D) Maximum-likelihood trees for HA (B), PB2 (C), and M (D) genes with a sample of BLAST-matched sequences (https://blast.ncbi.nlm.nih.gov). Bootstrap values >90% for key nodes are shown. Scale bars indicate number of nucleotide substitutions per site for each gene. HA, hemagglutinin; LPAI, low pathogenicity avian influenza; M, matrix protein; NP, nucleoprotein; NS, nonstructural; PA, polymerase acidic; PB1, polymerase basic 1; PB2, polymerase basic 2.
1These authors contributed equally to this article.