Volume 31, Number 1—January 2025
Dispatch
Clonal Complex 398 Methicillin-Resistant Staphylococcus aureus Producing Panton-Valentine Leukocidin, Czech Republic, 2023
Figure
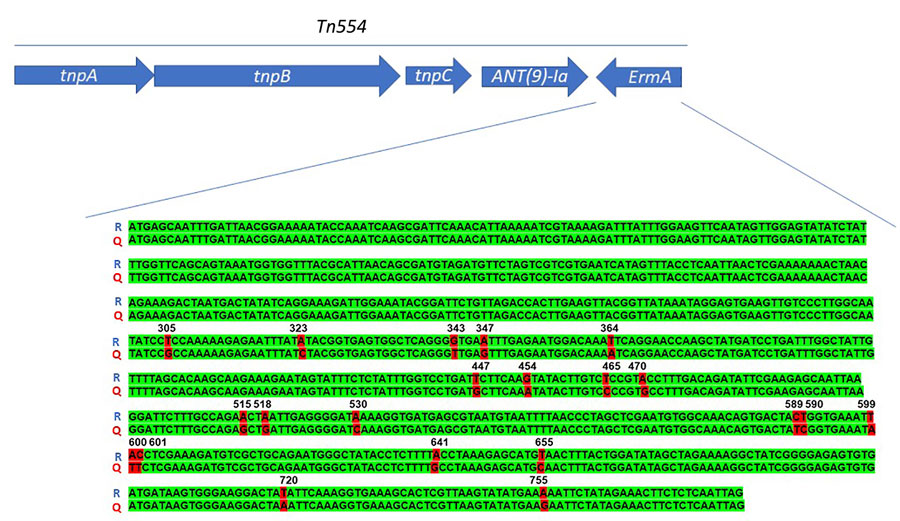
Figure. Schematic representation of the Tn544 transposon carrying the ermA and ant(9)-Ia genes representative of methicillin-resistant Staphylococcus aureus Panton-Valentine–positive clonal complex 398. The ermA gene sequence of the strains isolated from patients in the Czech Republic in 2023 align with the reference sequence of the ResFinder software (Genomic Epidemiology Center, http://www.genomicepidemiology.org). The resistance genes ermA and ant(9)-Ia (arrows) are transcribed in different directions because they are located on different strands of the DNA. Q, query sequence; R, reference sequence; tnpA, transposase A; tnpB, transposase B; tnpC, transposase C.
References
- Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, et al. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio. 2012;3:e00305–11. DOIPubMedGoogle Scholar
- Yu F, Chen Z, Liu C, Zhang X, Lin X, Chi S, et al. Prevalence of Staphylococcus aureus carrying Panton-Valentine leukocidin genes among isolates from hospitalised patients in China. Clin Microbiol Infect. 2008;14:381–4. DOIPubMedGoogle Scholar
- Welinder-Olsson C, Florén-Johansson K, Larsson L, Oberg S, Karlsson L, Ahrén C. Infection with Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus t034. Emerg Infect Dis. 2008;14:1271–2. DOIPubMedGoogle Scholar
- Gooskens J, Konstantinovski MM, Kraakman MEM, Kalpoe JS, van Burgel ND, Claas ECJ, et al. Panton-Valentine leukocidin-positive CC398 MRSA in urban clinical settings, the Netherlands. Emerg Infect Dis. 2023;29:1055–7. DOIPubMedGoogle Scholar
- Møller JK, Larsen AR, Østergaard C, Møller CH, Kristensen MA, Larsen J. International travel as source of a hospital outbreak with an unusual meticillin-resistant Staphylococcus aureus clonal complex 398, Denmark, 2016. Euro Surveill. 2019;24:
1800680 . DOIPubMedGoogle Scholar - Martineau F, Picard FJ, Roy PH, Ouellette M, Bergeron MG. Species-specific and ubiquitous DNA-based assays for rapid identification of Staphylococcus epidermidis. J Clin Microbiol. 1996;34:2888–93. DOIPubMedGoogle Scholar
- Boşgelmez-Tinaz G, Ulusoy S, Aridoğan B, Coşkun-Ari F. Evaluation of different methods to detect oxacillin resistance in Staphylococcus aureus and their clinical laboratory utility. Eur J Clin Microbiol Infect Dis. 2006;25:410–2. DOIPubMedGoogle Scholar
- Lina G, Piémont Y, Godail-Gamot F, Bes M, Peter MO, Gauduchon V, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–32. DOIPubMedGoogle Scholar
- Stegger M, Lindsay JA, Moodley A, Skov R, Broens EM, Guardabassi L. Rapid PCR detection of Staphylococcus aureus clonal complex 398 by targeting the restriction-modification system carrying sau1-hsdS1. J Clin Microbiol. 2011;49:732–4. DOIPubMedGoogle Scholar
- European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters version 14.0, valid from 2024-01-01 [cited 2024 Mar 11]. https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_14.0_Breakpoint_Tables.pdf
- Coombs GW, Daley D, Shoby P, Yee NWT, Robinson JO, Murray R, et al. Genomic characterisation of CC398 MRSA causing severe disease in Australia. Int J Antimicrob Agents. 2022;59:
106577 . DOIPubMedGoogle Scholar - Schouls LM, Witteveen S, van Santen-Verheuvel M, de Haan A, Landman F, van der Heide H, et al.; Dutch MRSA surveillance study group. Molecular characterization of MRSA collected during national surveillance between 2008 and 2019 in the Netherlands. Commun Med (Lond). 2023;3:123. DOIPubMedGoogle Scholar
- Liang B, Mai J, Liu Y, Huang Y, Zhong H, Xie Y, et al. Prevalence and characterization of Staphylococcus aureus isolated from women and children in Guangzhou, China. Front Microbiol. 2018;9:2790. DOIPubMedGoogle Scholar
- Koike S, Aminov RI, Yannarell AC, Gans HD, Krapac IG, Chee-Sanford JC, et al. Molecular ecology of macrolide-lincosamide-streptogramin B methylases in waste lagoons and subsurface waters associated with swine production. Microb Ecol. 2010;59:487–98. DOIPubMedGoogle Scholar
Page created: December 02, 2024
Page updated: December 22, 2024
Page reviewed: December 22, 2024
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.