Volume 31, Number 10—October 2025
Dispatch
Genomic Investigation of Disseminated Gonococcal Infections, Minnesota, USA, 2024
Figure 3
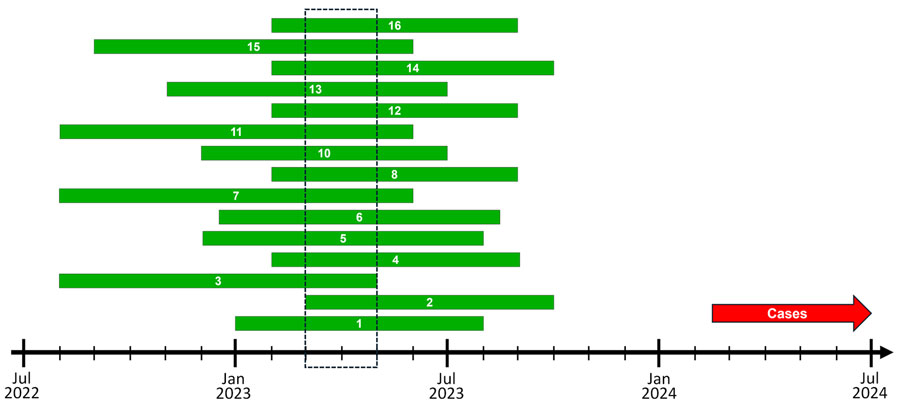
Figure 3. Phylodynamic inference of the emergence of Neisseria gonorrhoeae sequence type (ST) 11184 genomes that infected disseminated gonococcal infection cases, Minnesota, USA. The green bars denote the 90% CIs of the estimated time to most recent common ancestor (tMRCA) of a clade of 18 ST11184 genomes from Minnesota, rounded to the nearest month. We calculated tMRCAs by refining a maximum-likelihood, distance-scaled, core gene phylogenetic tree of publicly available ST11184 genomes into a time-scaled tree by using TreeTime v0.11.4 software (Appendix) (10). The dashed box denotes the period in which the tMRCA 90% CIs of all 16 iterations of time-scaled refinement overlapped. The red arrow denotes the time after the earliest collection of an isolate from a 2024 disseminated gonococcal infection case in Minnesota.
References
- Marrazzo JM, Apicella MA. Neisseria gonorrhoeae (gonorrhea). In: Blaser MJ, Cohen JI, Holland SM, editors. Mandell, Douglas, and Bennett’s principles and practice of infectious diseases. Amsterdam: Elsevier; 2015. p. 2446–2462
- Cartee JC, Joseph SJ, Weston E, Pham CD, Thomas JC IV, Schlanger K, et al. Phylogenomic comparison of Neisseria gonorrhoeae causing disseminated gonococcal infections and uncomplicated gonorrhea in Georgia, United States. Open Forum Infect Dis. 2022;9:ofac247. DOIGoogle Scholar
- Wisconsin State Laboratory of Hygiene. Spriggan, 2022 [cited 2025 May 27]. https://github.com/wslh-bio/spriggan
- Wisconsin State Laboratory of Hygiene. Dryad, 2023 [cited 2025 May 27]. https://github.com/wslh-bio/dryad
- Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. DOIPubMedGoogle Scholar
- Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. DOIPubMedGoogle Scholar
- Golparian D, Sánchez-Busó L, Cole M, Unemo M. Neisseria gonorrhoeae Sequence Typing for Antimicrobial Resistance (NG-STAR) clonal complexes are consistent with genomic phylogeny and provide simple nomenclature, rapid visualization and antimicrobial resistance (AMR) lineage predictions. J Antimicrob Chemother. 2021;76:940–4. DOIPubMedGoogle Scholar
- Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom. 2021;7:
000685 . DOIPubMedGoogle Scholar - Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, et al. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol. 2020;21:180. DOIPubMedGoogle Scholar
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic interence in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Sagulenko P, Puller V, Neher RA. TreeTime: maximum-likelihood phylodynamic analysis. Virus Evol. 2018;4:vex042. DOIGoogle Scholar
- Seemann T, Klotzl F, Page AJ. snp-dists. Pairwise SNP distance matrix from a FASTA sequence alignment. 2018 [cited 2025 May 27]. https://github. com/tseemann/snp-dists
- Welch G, Reed GW, Rice PA, Ram S. A meta-analysis to quantify the risk of disseminated gonococcal infection with Porin B serotype. Open Forum Infect Dis. 2024;11:ofae389. DOIGoogle Scholar
- Shaskolskiy B, Kravtsov D, Kandinov I, Gorshkova S, Kubanov A, Solomka V, et al. Comparative whole-genome analysis of Neisseria gonorrhoeae isolates revealed changes in the gonococcal genetic island and specific genes as a link to antimicrobial resistance. Front Cell Infect Microbiol. 2022;12:
831336 . DOIPubMedGoogle Scholar - Reimche JL, Clemons AA, Chivukula VL, Joseph SJ, Schmerer MW, Pham CD, et al.; Antimicrobial-Resistant Working Group. Genomic analysis of 1710 surveillance-based Neisseria gonorrhoeae isolates from the USA in 2019 identifies predominant strain types and chromosomal antimicrobial-resistance determinants. Microb Genom. 2023;9:mgen001006. DOIGoogle Scholar