Volume 31, Supplement—May 2025
SUPPLEMENT ISSUE
Supplement
Real-Time Use of Monkeypox Virus Genomic Surveillance, King County, Washington, USA, 2022–2024
Figure
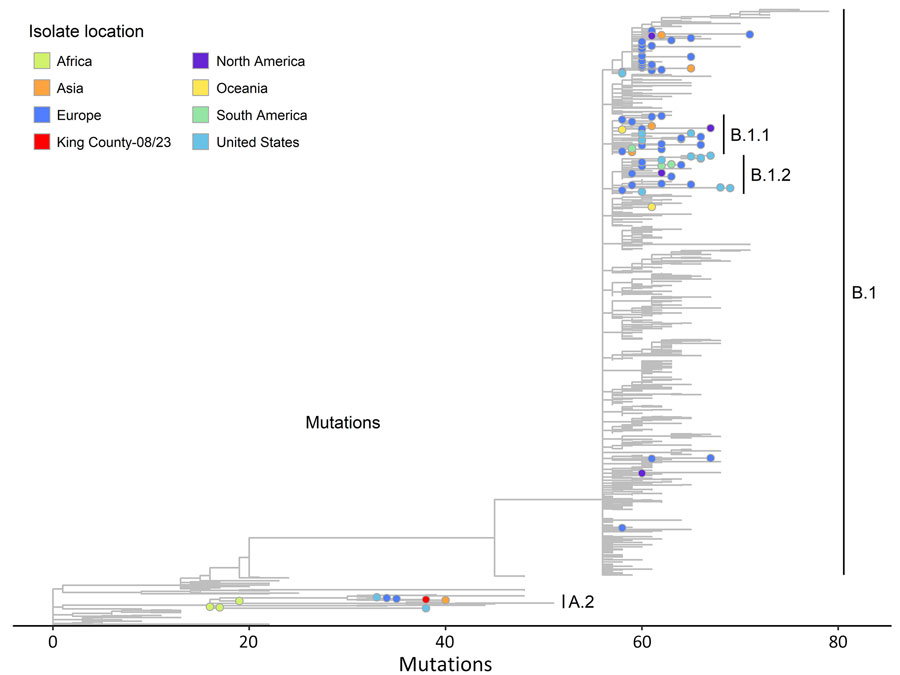
Figure. Phylogenetic reference tree of clade IIb monkeypox virus sequences as of December 2023 from study of real-time use of monkeypox virus genomic surveillance, King County, Washington, USA, 2022–2024. The tree shows the viral strain of sublineage A.2.1 identified from a King County, resident in fall 2023 (in red) is highly diverged from the B.1 lineage. Colors correspond to the location of the case from which a viral isolate was sampled and are either a region, the United States, or King County, for the specific isolate of interest in this case report. Phylogenetic tree generated using Nextclade dataset for “Mpox virus (All Clades)” in Auspice v2.61.1 (https://github.com/nextstrain/auspice.us). The reference sequence used in the tree is the clade IIb GenBank reference sequence (accession no. NC_063383.1). Data sourced from GenBank on December 8, 2023 (13). Sequences were downsampled by the Nextstrain team to ≈500 sequences with the goal of capturing monkeypox virus diversity across geography, collection dates, and lineages. The dataset is archived by Nextclade (https://github.com/nextstrain/Nextclade_data/tree/master/data_output/nextstrain/mpox/all-clades/2024-01-16--20-31-02Z). The figure is filtered to the clade IIb branch within the larger dataset. Branches are shown for all lineages within clade IIb, and specific nodes are shown for selected lineages A.2, A.2.1, B.1.1, B.1.2, and B.1.3. A table of the nodes with metadata is included in the Appendix.
References
- Laurenson-Schafer H, Sklenovská N, Hoxha A, Kerr SM, Ndumbi P, Fitzner J, et al.; WHO mpox Surveillance and Analytics team. Description of the first global outbreak of mpox: an analysis of global surveillance data. Lancet Glob Health. 2023;11:e1012–23. DOIPubMedGoogle Scholar
- Roychoudhury P, Sereewit J, Xie H, Nunley E, Bakhash SM, Lieberman NAP, et al. Genomic analysis of early monkeypox virus outbreak strains, Washington, USA. Emerg Infect Dis. 2023;29:644–6. DOIPubMedGoogle Scholar
- Genomic epidemiology of monkeypox virus – Washington state focused build. [cited 2024 Nov 19] https://nextstrain.org/groups/waphl/wa/hmpxv1
- Hadfield J, Megill C, Bell SM, Huddleston J, Potter B, Callender C, et al. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. 2018;34:4121–3. DOIPubMedGoogle Scholar
- Aksamentov I, Roemer C, Hodcroft EB, Neher RA. Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw. 2021;6:3773. DOIGoogle Scholar
- Gigante CM, Korber B, Seabolt MH, Wilkins K, Davidson W, Rao AK, et al. Multiple lineages of monkeypox virus detected in the United States, 2021-2022. Science. 2022;378:560–5. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Mpox in the U.S. 2024. [cited 2024 Nov 19] https://www.cdc.gov/poxvirus/mpox/symptoms/index.html.
- Gehre F, Lagu HI, Achol E, Omari N, Ochido G, Shand K, et al. The East African Community mobile laboratory network prepares for monkeypox outbreaks. J Public Health Africa. 2023;14:2309. DOIPubMedGoogle Scholar
- Hermez J, El Helou R, Sawaya T, Sader G, Jamil MS, Alaama AS, et al. Emergence of mpox in the Eastern Mediterranean Region: Data assessment and implications for a public health response. J Infect Public Health. 2024;17:
102565 . DOIPubMedGoogle Scholar - Dung NT, Hung LM, Hoa HTT, Nga LH, Hong NTT, Thuong TC, et al. Monkeypox virus infection in 2 female travelers returning to Vietnam from Dubai, United Arab Emirates, 2022. Emerg Infect Dis. 2023;29:778–81. DOIPubMedGoogle Scholar
- Shete AM, Yadav PD, Kumar A, Patil S, Patil DY, Joshi Y, et al. Genome characterization of monkeypox cases detected in India: Identification of three sub clusters among A.2 lineage. J Infect. 2023;86:66–117. DOIPubMedGoogle Scholar
- El Dine FB, Gebreal A, Samhouri D, Estifanos H, Kourampi I, Abdelrhem H, et al. Ethical considerations during Mpox Outbreak: a scoping review. BMC Med Ethics. 2024;25:79. DOIPubMedGoogle Scholar
- Sayers EW, Cavanaugh M, Clark K, Pruitt KD, Sherry ST, Yankie L, et al. GenBank 2024 Update. Nucleic Acids Res. 2024;52(D1):D134–7. DOIPubMedGoogle Scholar