Dengue and Other Arbovirus Infections among Schoolchildren, Haiti, 2021
Rigan Louis, Tracey L. Moquin, Carla Mavian, Assonic Barthelemy, Ruiyu Pu, Benjamin Anderson, V. Madsen Beau De Rochars, Maureen T. Long, Marco Salemi, John A. Lednicky, and J. Glenn Morris

Author affiliation: University of Florida College of Nursing, Gainesville, Florida, USA (R. Louis); University of Florida Emerging Pathogens Institute, Gainesville (R. Louis, T.L. Moquin, C. Mavian, R. Pu, B. Anderson, V.M.B. De Rochars, M.T. Long, M. Salemi, J.A. Lednicky, J.G. Morris, Jr.); University of Florida College of Public Health and Health Professions, Gainesville (T.L. Moquin, B. Anderson, V.M.B. De Rochars, J A. Lednicky); Smithsonian's National Zoo and Conservation Biology Institute, Washington, DC, USA (C. Mavian); University of Florida College of Medicine, Gainesville (C. Mavian, M. Salemi); Love A Child Foundation Medical Clinic, Fond Parisien, Haiti (A. Barthelemy)
Main Article
Figure 4
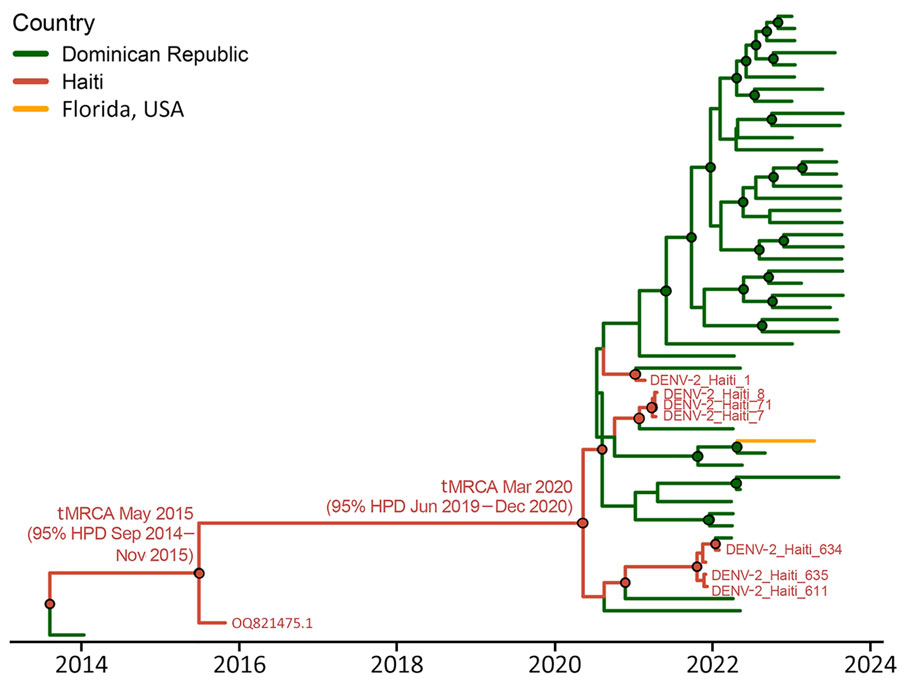
Figure 4. Phylogeography for the American/Asian genotype of DENV containing new Haiti isolates. Time-scaled phylogenetic maximum clade creditability tree for Haiti clade 2 (including GenBank accession no. OQ821475.1, isolated in 2015, and new 2021 strains, shown in red) was inferred using the phylogeographic frameworks in BEAST version 1.10.4 (https://beast.community) and enforcing the Bayesian Skyline demographic prior with an uncorrelated lognormal relaxed clock. Branch colors represent country of origin of the genome, and posterior probability bootstrap support >0.90 at each node is shown with a circle colored by the ancestral country of origin. DENV, dengue virus; HPD, highest posterior density; tMRCA, time to most recent common ancestor.
Main Article
Page created: December 30, 2024
Page updated: January 31, 2025
Page reviewed: January 31, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
