Volume 31, Number 2—February 2025
Dispatch
Seoul Virus Infection and Subsequent Guillain-Barré Syndrome in Traveler Returning to France from Kenya, 2022
Figure
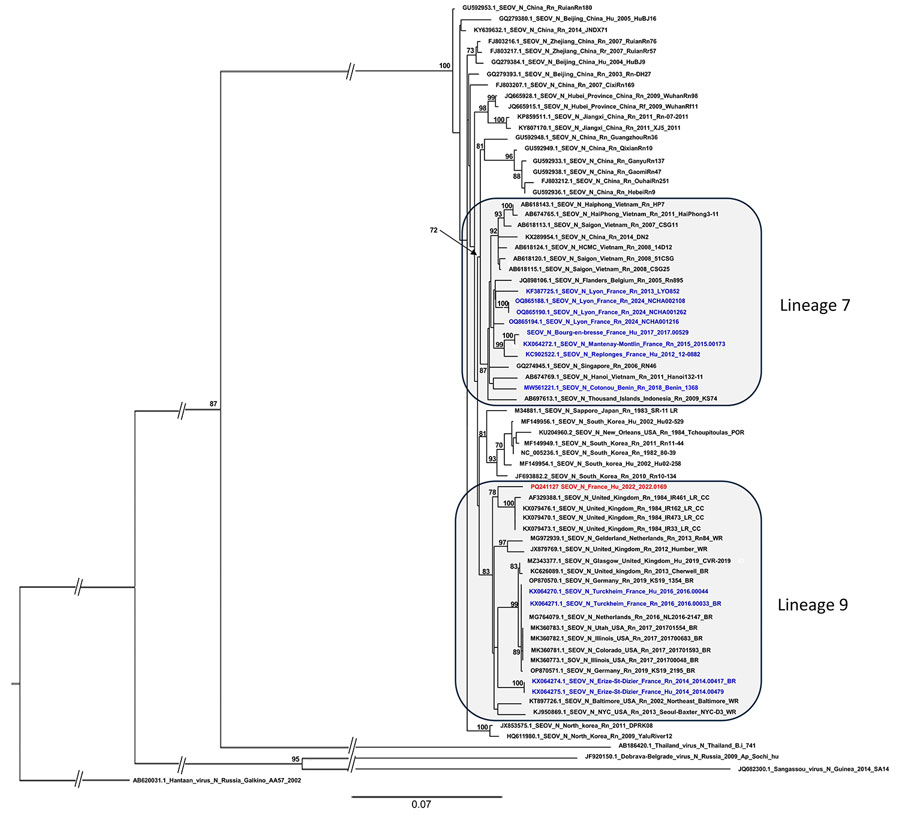
Figure. Phylogenetic tree based on the complete coding region (1,290 bp) of the small segment of SEOV strain detected in an infected patient in France, 2022, and representative strains of SEOV and other hantavirus species. The complete segment Bayesian tree was reconstructed using MAFFT version 7.023b (https://mafft.cbrc.jp/alignment/software) and RAxML 8.2 (https://cme.h-its.org/exelixis/web/software/raxml) with the general time-reversible plus gamma distribution substitution model and a rapid bootstrap (i.e., general time-reversible invariable site plus discrete Gamma model, bootstraps = 1,000). The numbers at each node are bootstrap probabilities (>70%) as determined for 1,000 iterations. The SEOV strain Hu_2022_2022.0169 (GenBank accession no. PQ241127) retrieved in this study is indicated in red, whereas other sequences from France and Benin are represented in blue. GenBank accession numbers are provided for reference viruses. Hantaan virus was used as outgroup. Scale bars indicate nucleotide substitutions per site. BR, breeder rat (includes feeder and pet rats); CC, cell culture; Hu, human; LR, laboratory rat; N, nucleoprotein; Rn, Rattus norvegicus; Rr, R. rattus; SEOV, Seoul virus; WR, wild rat.