Volume 31, Number 3—March 2025
Synopsis
Genetic Diversity and Geographic Spread of Henipaviruses
Figure 1
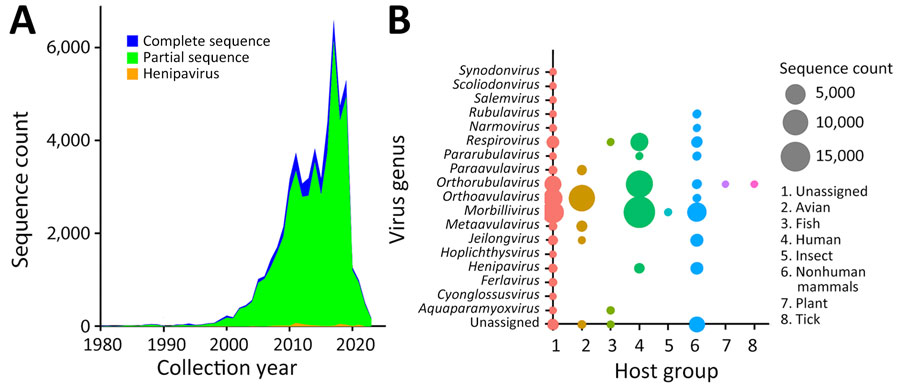
Figure 1. Trend in paramyxovirus sequences submitted to the National Center for Biotechnology Information Virus database (https://www.ncbi.nlm.nih.gov/labs/virus), 1980–2023. A) Sequence count by collection year, showing all complete and partial sequences compared with all henipaviruses. B) Virus genera and sequence counts by major host group from the VIRION database (32).
References
- Wang L, Harcourt BH, Yu M, Tamin A, Rota PA, Bellini WJ, et al. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 2001;3:279–87. DOIPubMedGoogle Scholar
- Quarleri J, Galvan V, Delpino MV. Henipaviruses: an expanding global public health concern? Geroscience. 2022;44:2447–59. DOIPubMedGoogle Scholar
- Aljofan M. Hendra and Nipah infection: emerging paramyxoviruses. Virus Res. 2013;177:119–26. DOIPubMedGoogle Scholar
- Gazal S, Sharma N, Gazal S, Tikoo M, Shikha D, Badroo GA, et al. Nipah and Hendra viruses: deadly zoonotic paramyxoviruses with the potential to cause the next pandemic. Pathogens. 2022;11:1419. DOIPubMedGoogle Scholar
- Breed AC, Meers J, Sendow I, Bossart KN, Barr JA, Smith I, et al. The distribution of henipaviruses in Southeast Asia and Australasia: is Wallace’s line a barrier to Nipah virus? PLoS One. 2013;8:
e61316 . DOIPubMedGoogle Scholar - Chen JM, Yu M, Morrissy C, Zhao YG, Meehan G, Sun YX, et al. A comparative indirect ELISA for the detection of henipavirus antibodies based on a recombinant nucleocapsid protein expressed in Escherichia coli. J Virol Methods. 2006;136:273–6. DOIPubMedGoogle Scholar
- Parashar UD, Sunn LM, Ong F, Mounts AW, Arif MT, Ksiazek TG, et al. Case-control study of risk factors for human infection with a new zoonotic paramyxovirus, Nipah virus, during a 1998-1999 outbreak of severe encephalitis in Malaysia. J Infect Dis. 2000;181:1755–9. DOIPubMedGoogle Scholar
- Luby SP, Gurley ES, Hossain MJ. Transmission of human infection with Nipah virus. Clin Infect Dis. 2009;49:1743–8. DOIPubMedGoogle Scholar
- Taylor J, Thompson K, Annand EJ, Massey PD, Bennett J, Eden JS, et al. Novel variant Hendra virus genotype 2 infection in a horse in the greater Newcastle region, New South Wales, Australia. One Health. 2022;15:
100423 . DOIPubMedGoogle Scholar - Smith C, Skelly C, Kung N, Roberts B, Field H. Flying-fox species density—a spatial risk factor for Hendra virus infection in horses in eastern Australia. PLoS One. 2014;9:
e99965 . DOIPubMedGoogle Scholar - Edson D, Peel AJ, Huth L, Mayer DG, Vidgen ME, McMichael L, et al. Time of year, age class and body condition predict Hendra virus infection in Australian black flying foxes (Pteropus alecto). Epidemiol Infect. 2019;147:
e240 . DOIPubMedGoogle Scholar - Ching PKG, de los Reyes VC, Sucaldito MN, Tayag E, Columna-Vingno AB, Malbas FF Jr, et al. Outbreak of henipavirus infection, Philippines, 2014. Emerg Infect Dis. 2015;21:328–31. DOIPubMedGoogle Scholar
- Islam A, Cannon DL, Rahman MZ, Khan SU, Epstein JH, Daszak P, et al. Nipah virus exposure in domestic and peridomestic animals living in human outbreak sites, Bangladesh, 2013–2015. Emerg Infect Dis. 2023;29:393–6. DOIPubMedGoogle Scholar
- Becker DJ, Crowley DE, Washburne AD, Plowright RK. Temporal and spatial limitations in global surveillance for bat filoviruses and henipaviruses. Biol Lett. 2019;15:
20190423 . DOIPubMedGoogle Scholar - Li Y, Wang J, Hickey AC, Zhang Y, Li Y, Wu Y, et al. Antibodies to Nipah or Nipah-like viruses in bats, China. Emerg Infect Dis. 2008;14:1974–6. DOIPubMedGoogle Scholar
- Hayman DTS, Suu-Ire R, Breed AC, McEachern JA, Wang L, Wood JLN, et al. Evidence of henipavirus infection in West African fruit bats. PLoS One. 2008;3:
e2739 . DOIPubMedGoogle Scholar - Madera S, Kistler A, Ranaivoson HC, Ahyong V, Andrianiaina A, Andry S, et al. Discovery and genomic characterization of a novel henipavirus, Angavokely virus, from fruit bats in Madagascar. J Virol. 2022;96:
e0092122 . DOIPubMedGoogle Scholar - Hernández LHA, da Paz TYB, Silva SPD, Silva FSD, Barros BCV, Nunes BTD, et al. First genomic evidence of a henipa-like virus in Brazil. Viruses. 2022;14:2167. DOIPubMedGoogle Scholar
- Horemans M, Van Bets J, Joly Maes T, Maes P, Vanmechelen B. Discovery and genome characterization of six new orthoparamyxoviruses in small Belgian mammals. Virus Evol. 2023;9:
vead065 . DOIPubMedGoogle Scholar - Zhang X-A, Li H, Jiang F-C, Zhu F, Zhang Y-F, Chen J-J, et al. A zoonotic henipavirus in febrile patients in China. N Engl J Med. 2022;387:470–2. DOIPubMedGoogle Scholar
- Lee SH, Kim K, Kim J, No JS, Park K, Budhathoki S, et al. Discovery and genetic characterization of novel paramyxoviruses related to the genus Henipavirus in Crocidura species in the Republic of Korea. Viruses. 2021;13:2020. DOIPubMedGoogle Scholar
- Chakraborty S, Chandran D, Mohapatra RK, Islam MA, Alagawany M, Bhattacharya M, et al. Langya virus, a newly identified Henipavirus in China - Zoonotic pathogen causing febrile illness in humans, and its health concerns: Current knowledge and counteracting strategies - Correspondence. Int J Surg. 2022;105:
106882 . DOIPubMedGoogle Scholar - Weatherman S, Feldmann H, de Wit E. Transmission of henipaviruses. Curr Opin Virol. 2018;28:7–11. DOIPubMedGoogle Scholar
- Wu Z, Yang L, Yang F, Ren X, Jiang J, Dong J, et al. Novel henipa-like virus, Mojiang paramyxovirus, in rats, China, 2012. Emerg Infect Dis. 2014;20:1064–6. DOIPubMedGoogle Scholar
- Hayman DTS, Wang LF, Barr J, Baker KS, Suu-Ire R, Broder CC, et al. Antibodies to henipavirus or henipa-like viruses in domestic pigs in Ghana, West Africa. PLoS One. 2011;6:
e25256 . DOIPubMedGoogle Scholar - Baker KS, Leggett RM, Bexfield NH, Alston M, Daly G, Todd S, et al. Metagenomic study of the viruses of African straw-coloured fruit bats: detection of a chiropteran poxvirus and isolation of a novel adenovirus. Virology. 2013;441:95–106. DOIPubMedGoogle Scholar
- Lu M, Yao Y, Zhang X, Liu H, Gao G, Peng Y, et al. Both chimpanzee adenovirus-vectored and DNA vaccines induced long-term immunity against Nipah virus infection. NPJ Vaccines. 2023;8:170. DOIPubMedGoogle Scholar
- Mire CE, Geisbert JB, Agans KN, Feng YR, Fenton KA, Bossart KN, et al. A recombinant Hendra virus G glycoprotein subunit vaccine protects nonhuman primates against Hendra virus challenge. J Virol. 2014;88:4624–31. DOIPubMedGoogle Scholar
- Huang X, Li Y, Li R, Wang S, Yang L, Wang S, et al. Nipah virus attachment glycoprotein ectodomain delivered by type 5 adenovirus vector elicits broad immune response against NiV and HeV. Front Cell Infect Microbiol. 2023;13:
1180344 . DOIPubMedGoogle Scholar - Naveed M, Mehmood S, Aziz T, Hammad Arif M, Ali U, Nouroz F, et al. An mRNA-based reverse-vaccinology strategy to stimulate the immune response against Nipah virus in humans using fusion glycoproteins. Acta Biochim Pol. 2023;70:623–31. DOIPubMedGoogle Scholar
- Broder CC, Xu K, Nikolov DB, Zhu Z, Dimitrov DS, Middleton D, et al. A treatment for and vaccine against the deadly Hendra and Nipah viruses. Antiviral Res. 2013;100:8–13. DOIPubMedGoogle Scholar
- Carlson CJ, Gibb RJ, Albery GF, Brierley L, Connor RP, Dallas TA, et al. The global virome in one Network (VIRION): an atlas of vertebrate-virus associations. MBio. 2022;13:
e0298521 . DOIPubMedGoogle Scholar - Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. DOIPubMedGoogle Scholar
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–3. DOIPubMedGoogle Scholar
- Shen W, Sipos B, Zhao L. SeqKit2: A Swiss army knife for sequence and alignment processing. iMeta. 2024;3:
e191 . DOIPubMedGoogle Scholar - Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, et al. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–4. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Kumar S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol Biol Evol. 2021;38:3022–7. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Hasegawa M, Kishino H, Yano T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–74. DOIPubMedGoogle Scholar
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol. 2018;67:901–4. DOIPubMedGoogle Scholar
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–9. DOIPubMedGoogle Scholar
- Crowley D, Becker D, Washburne A, Plowright R. Identifying suspect bat reservoirs of emerging infections. Vaccines (Basel). 2020;8:228. DOIPubMedGoogle Scholar
- Gu SH, Nicolas V, Lalis A, Sathirapongsasuti N, Yanagihara R. Complete genome sequence and molecular phylogeny of a newfound hantavirus harbored by the Doucet’s musk shrew (Crocidura douceti) in Guinea. Infect Genet Evol. 2013;20:118–23. DOIPubMedGoogle Scholar
- Mortlock M, Geldenhuys M, Dietrich M, Epstein JH, Weyer J, Pawęska JT, et al. Seasonal shedding patterns of diverse henipavirus-related paramyxoviruses in Egyptian rousette bats. Sci Rep. 2021;11:24262. DOIPubMedGoogle Scholar
- Isaacs A, Low YS, Macauslane KLL, Seitanidou J, Pegg CLL, Cheung STM, et al. Structure and antigenicity of divergent Henipavirus fusion glycoproteins. Nat Commun. 2023;14:3577. DOIPubMedGoogle Scholar
Page created: February 04, 2025
Page updated: February 28, 2025
Page reviewed: February 28, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.