Volume 31, Number 4—April 2025
Dispatch
Yaws Circulating in Nonhuman Primates, Uganda and Rwanda
Figure 2
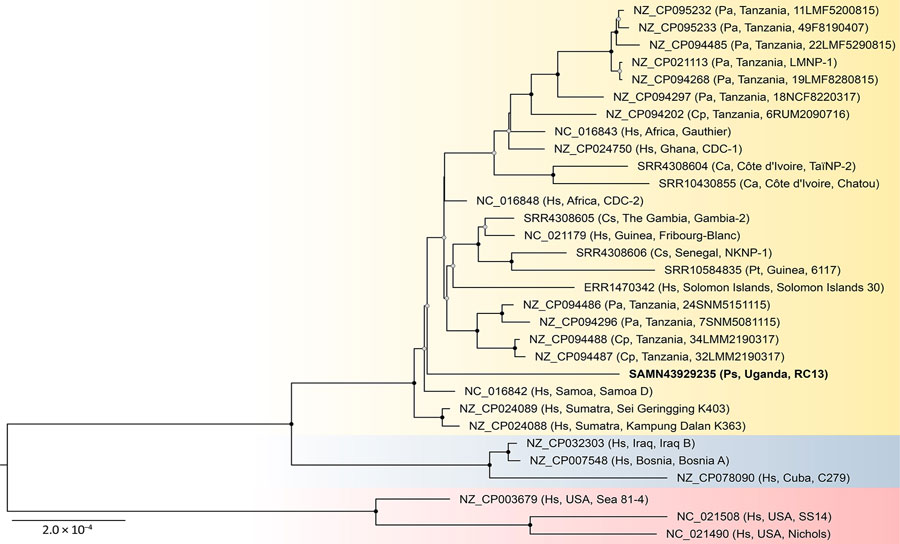
Figure 2. Maximum-likelihood phylogenetic tree of Treponema pallidum genomes from study of yaws circulating in nonhuman primates, Uganda and Rwanda. The tree shows relationships among T. p. pertenue genomes (yellow) and is rooted with T. p. endemicum (blue) and T. p. pallidum (red) genomes. Bold text indicates sequence generated in this study from a Ugandan red colobus monkey. Taxon names include GenBank accession numbers, followed in parentheses by primate host, location of origin, and isolate name. The tree was inferred from a 1,072,667-position cleaned nucleotide alignment of 31 nonredundant sequences available in GenBank having genome coverage >97% and containing 4,716 variable positions. Black dots on nodes indicate bootstrap values of 100%; gray dots indicate 75%–99% bootstrap values based on 1,000 bootstrap replicates; values <75% are not shown. Scale bar indicates nucleotide substitutions per site. Ca, Cercocebus atys; Cp, Chlorocebus pygerythrus; Cs, Cercocebus sabaeus; Hs, Homo sapiens; Pa, Papio anubis; Ps, Piliocolobus tephrosceles; Pt, Pan troglodytes.