Volume 31, Number 4—April 2025
Research Letter
Spread of Dual-Resistant Mycoplasma genitalium Clone among Men, France, 2021–2022
Figure
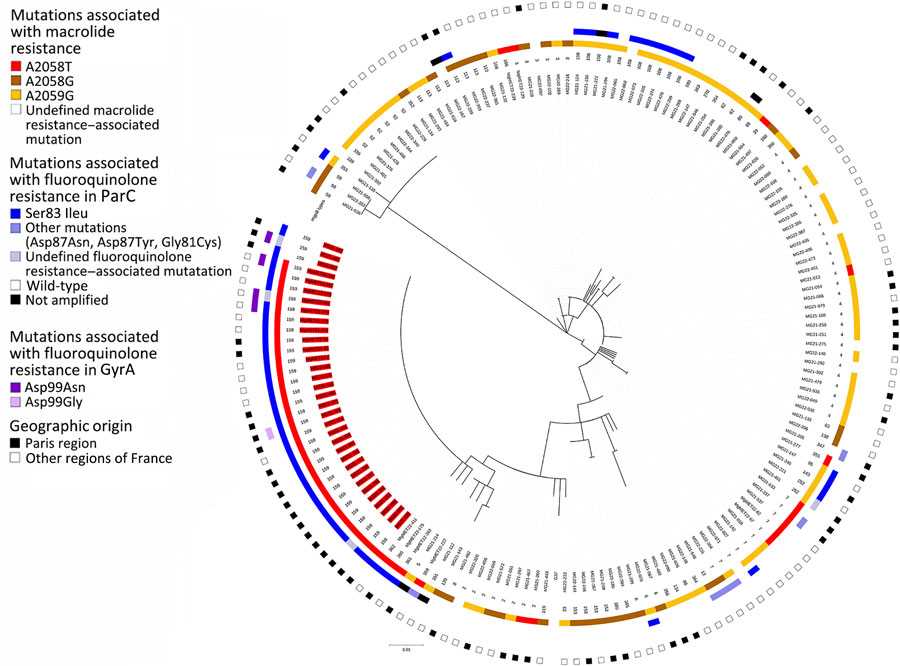
Figure. Maximum-likelihood tree in a study of spread of dual-resistant Mycoplasma genitalium clone among men, France, 2021–2022. The tree is based on the mgpB type of 163 successfully mgpB-typed M. genitalium–positive specimens harboring macrolide resistance–associated mutations. The tree was inferred using the maximum-likelihood method based on the Tamura 3–parameter model (T92 general plus invariable site model) constructed in MEGA7 (https://www.megasoftware.net). Branch support values were generated from 100 bootstrap replicates. The M. genitalium G37 strain (American Type Culture Collection no. 33530, https://www.atcc.org) sequence was used as a reference. The phylogenetic tree was annotated from the center to the periphery with the specimen name, the mgpB type, the macrolide resistance–associated mutation (Escherichia coli numbering), the fluoroquinolone resistance–associated ParC and GyrA mutations (M. genitalium numbering), and the geographic origin of the patient. Specimen names that were mgpB ST159 are highlighted in red. The phylogenetic tree was displayed and annotated using iTOL version 6 (https://itol.embl.de). Scale bar indicates the branch length corresponding to a genetic distance of 0.01, which indicates the average number of nucleotide substitutions per site.
1Members of the investigator group are listed at the end of this article.