Volume 31, Number 4—April 2025
Dispatch
Detection of Batborne Hantaviruses, Laos, 2023–2024
Figure 4
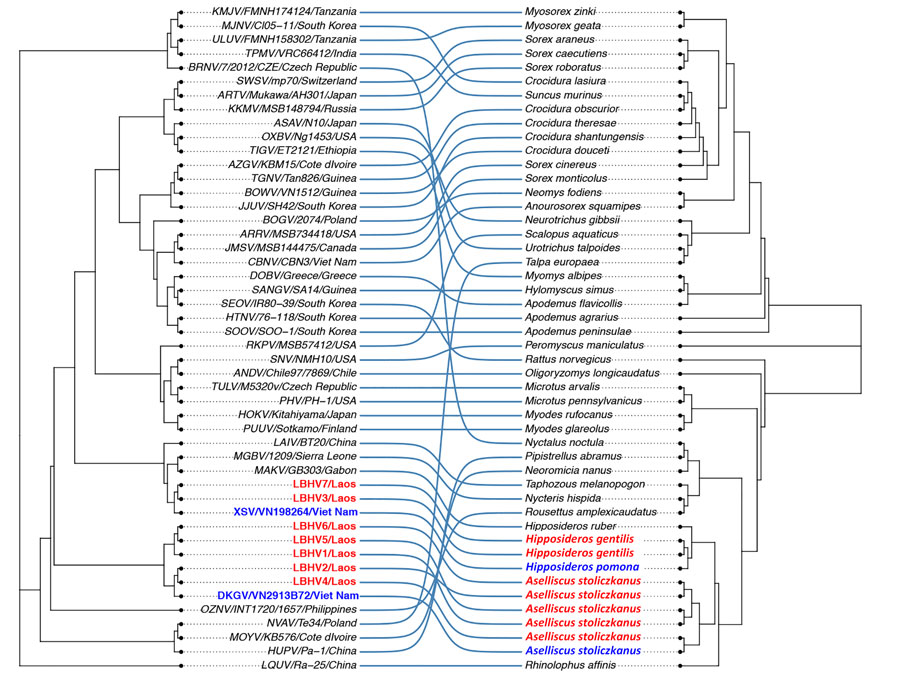
Figure 4. LBHVs and other mobatviruses from Southeast Asia showing detection of batborne hantaviruses, Laos, 2023–2024. The tanglegram compares maximum-likelihood trees from the hantavirus phylogeny in Figure 3 and the host cytochrome B gene. Red text indicates hantaviruses detected in Laos, and blue text indicate strains from Vietnam. The maximum-likelihood tree of the host cytochrome B gene was reconstructed with the same method as in Figure 3 with the AC = AT, CG = GT and equal base frequency plus proportion of invariable sites plus discrete Gamma model with default 4 rate categories model for cytochrome B, with 1,000 bootstrap replicates. LBHV, Lao batborne hantavirus. Additional virus abbreviations are given in the Appendix.
Page created: February 28, 2025
Page updated: March 24, 2025
Page reviewed: March 24, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.