Nipah Virus Detection in Pteropus hypomelanus Bats, Central Java, Indonesia
Dimas Bagus Wicaksono Putro, Arief Mulyono, Esti Rahardianingtyas, Aryo Ardanto, Arum Sih Joharina, Muhammad Choirul Hidajat, Yusnita Mirna Anggraeni, Ristiyanto Ristiyanto, Tika Fiona Sari, N.L.P. Indi Dharmayanti, Triwibowo Ambar Garjito, and R. Tedjo Sasmono

Author affiliation: Environmental Health National Referral Laboratory, Directorate General of Public Health, Ministry of Health of the Republic of Indonesia, Salatiga, Indonesia (D.B.W. Putro, E. Rahardianingtyas, A. Ardanto, A.S. Joharina, T.F. Sari); National Research and Innovation Agency, Bogor, Indonesia (A. Mulyono, M.C. Hidajat, Y.M. Anggraeni, Ristiyanto, N.L.P.I. Dharmayanti, T.A. Garjito, R.T. Sasmono)
Main Article
Figure 2
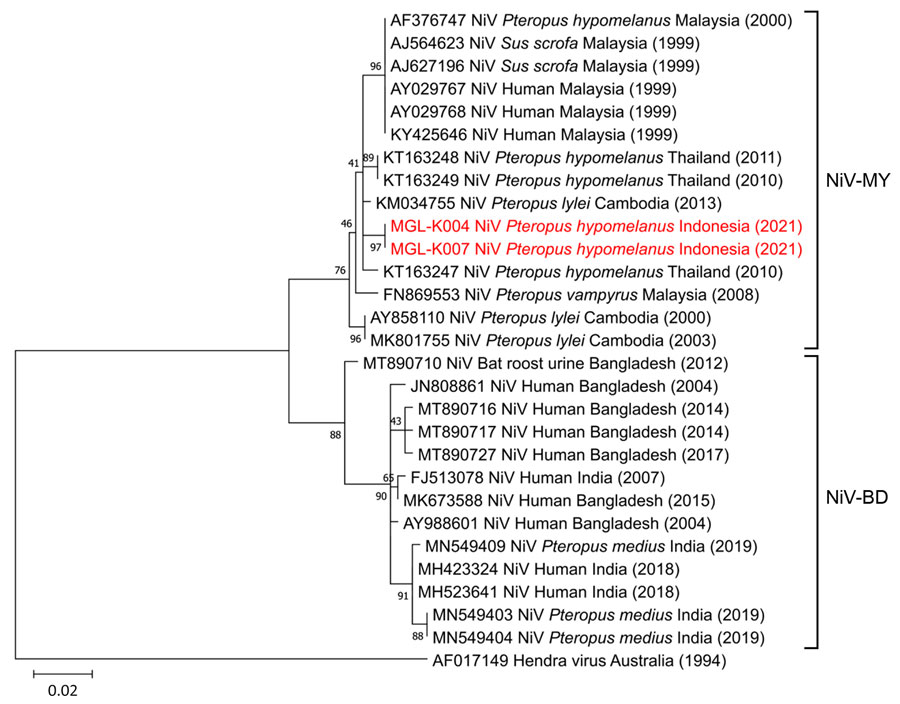
Figure 2. Phylogenetic tree showing NiV detection in Pteropus hypomelanus bats in Central Java, Indonesia. Tree illustrates evolutionary relationship of NiV isolates from Pteropus hypomelanus bats collected from animal markets in Central Java, Indonesia (specimen codes MGL-K004 and MGL-K007; red font) with strains from Southeast and South Asia (GenBank accession nos., host, and country of isolation indicated). Labels at right illustrates the 2 main genotypes of NiV. Hendra virus was used as an outgroup. The tree was constructed from 400-bp nucleocapsid gene sequences by using the maximum-likelihood algorithm performed in MEGA 11 software (http://www.megasoftware.net). Numbers to the left of the nodes are bootstrap percentages (1,000 replications). Scale bar indicates nucleotide substitutions per site. NiV, Nipah virus; NiV-BD, Bangladesh genotype; NiV-MY, Malaysia genotype.
Main Article
Page created: February 04, 2025
Page updated: March 24, 2025
Page reviewed: March 24, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
