Volume 31, Number 6—June 2025
Dispatch
Three Cases of Human Babesiosis, Italy, 2017–2020
Figure 2
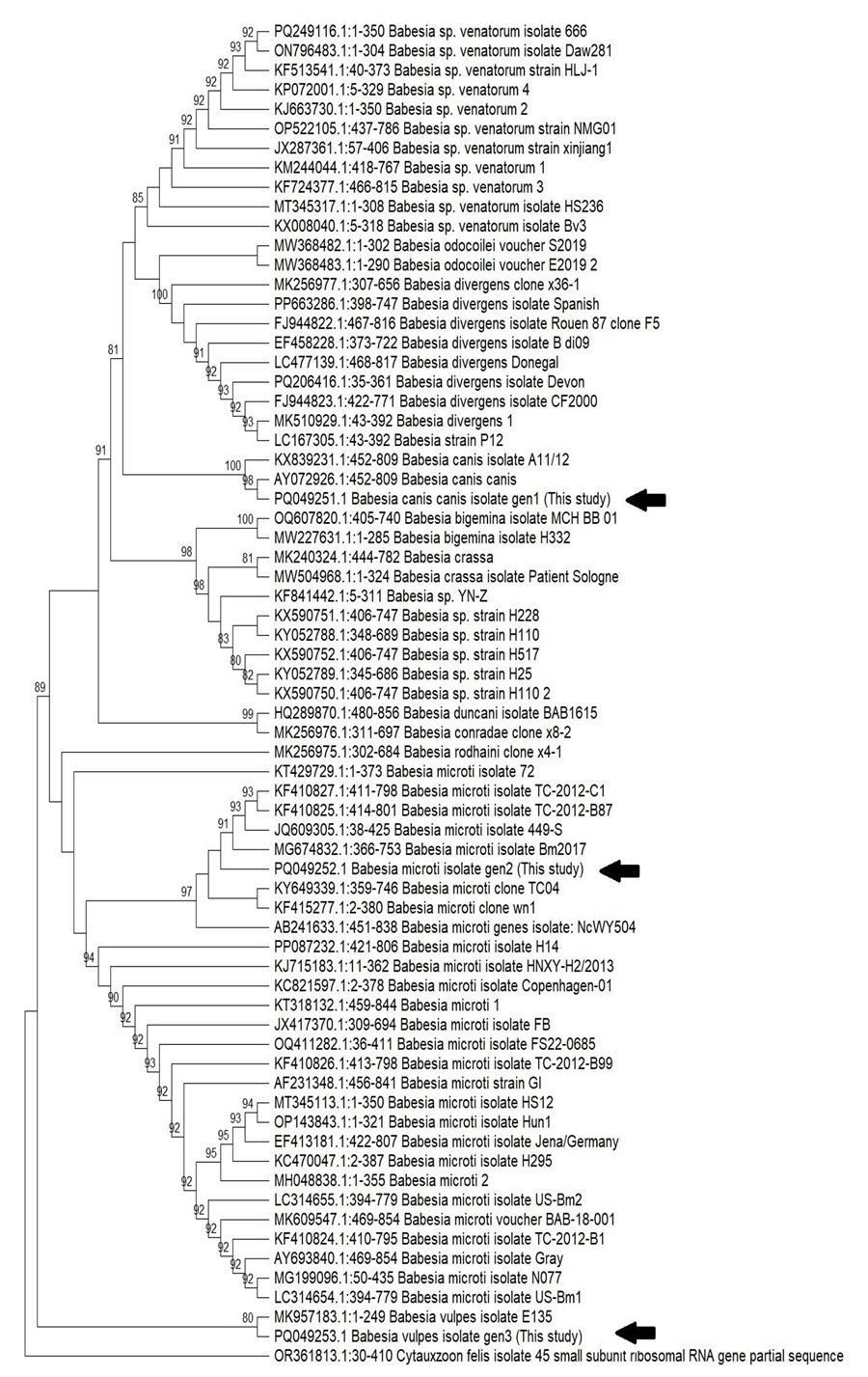
Figure 2. Phylogenetic analysis of the Babesia spp. in study of 3 cases of human babesiosis, Italy, 2017–2020. 18S ribosomal RNA gene sequences were amplified from the 3 clinical isolates from Italy. Arrows indicate the Babesia isolates from the 3 patients. Tree was inferred by using the maximum-likelihood method with 1,000 bootstrap replicates. Tree with the highest log likelihood (−3,487.14) is shown. Bootstrap values (cutoff at 80%) are indicated at specific branch nodes. 18S ribosomal RNA gene (partial) from Cytauxzoon felis protozoan parasite was used as the outlier. Tree not drawn to scale.
1These first authors contributed equally to this article.
Page created: April 23, 2025
Page updated: May 27, 2025
Page reviewed: May 27, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.