Volume 31, Number 6—June 2025
Dispatch
Highly Pathogenic Avian Influenza A(H5N1) in Wild Birds and a Human, British Columbia, Canada, 2024
Figure 3
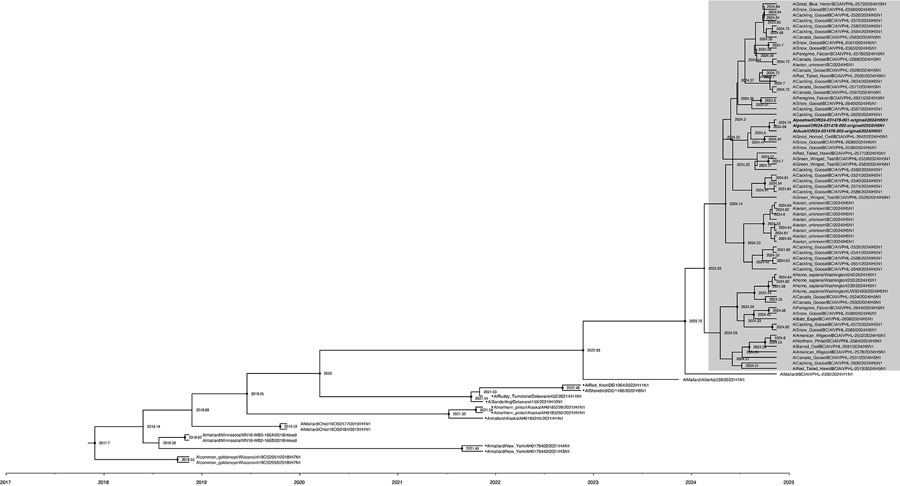
Figure 3. Phylogenetic relationships and possible origins of the neuraminidase (NA) segment (Am4N1) of the clade 2.3.4.4b D1.1 genotype detected in fall 2024 as part of a study of highly pathogenic avian influenza A(H5N1) in wild birds and a human, British Columbia, Canada, 2024. The maximum clade credibility tree was inferred by using BEAST v2.7.7 (6), incorporating NA gene sequences from D1.1 viruses and closely related sequences from wild birds detected in British Columbia and National Center for Biotechnology Information databases. Analysis used an uncorrelated relaxed log-normal clock, the Hasegawa-Kishino-Yano substitution model without gamma rate heterogeneity, and a coalescent Bayesian skyline tree prior. The posterior distribution was approximated using 100 million Markov chain Monte Carlo steps, sampled every 10,000 steps, with a 10% burn-in. All NA segments were identified as Am4N1 by GenoFLU except those indicated with a black dot on the tree tip label; non-Am4N1 sequences were classified as unassigned. Shaded box indicates NA segments from D1.1 and D1.2 H5N1 viruses. D1.2 samples are bolded and italicized. Node support values and branch lengths indicate the evolutionary divergence and probable reassortment timeframes leading to the emergence of the Am4N1 NA segment in the D1.1 genotype.
References
- Alkie TN, Lopes S, Hisanaga T, Xu W, Suderman M, Koziuk J, et al. A threat from both sides: Multiple introductions of genetically distinct H5 HPAI viruses into Canada via both East Asia-Australasia/Pacific and Atlantic flyways. Virus Evol. 2022;8:
veac077 . DOIPubMedGoogle Scholar - Youk S, Torchetti MK, Lantz K, Lenoch JB, Killian ML, Leyson C, et al. H5N1 highly pathogenic avian influenza clade 2.3.4.4b in wild and domestic birds: Introductions into the United States and reassortments, December 2021-April 2022. Virology. 2023;587:
109860 . DOIPubMedGoogle Scholar - Russell SL, Andrew CL, Yang KC, Coombe M, McGregor G, Redford T, et al. Descriptive epidemiology and phylogenetic analysis of highly pathogenic avian influenza H5N1 clade 2.3.4.4b in British Columbia (B.C.) and the Yukon, Canada, September 2022 to June 2023. Emerg Microbes Infect. 2024;13:
2392667 . DOIPubMedGoogle Scholar - Canadian Food Inspection Agency. Flocks in Canada where HPAI has been detected. 2024 November 28 [cited 2024 Nov 29]. https://inspection.canada.ca/en/animal-health/terrestrial-animals/diseases/reportable/avian-influenza/latest-bird-flu-situation/status-ongoing-response.
- Jassem AN, Roberts A, Tyson J, Zlosnik JEA, Russell SL, Caleta JM, et al. Critical illness in an adolescent with influenza A(H5N1) virus infection. N Engl J Med. 2025;392:927–9. DOIPubMedGoogle Scholar
- Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, et al. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLOS Comput Biol. 2019;15:
e1006650 . DOIPubMedGoogle Scholar - Signore AV, Joseph T, Ranadheera C, Erdelyan CNG, Alkie TN, Raj S, et al. Neuraminidase reassortment and oseltamivir resistance in clade 2.3.4.4b A(H5N1) viruses circulating among Canadian poultry, 2024. Emerg Microbes Infect. 2025;14:
2469643 . DOIPubMedGoogle Scholar - Animal and Plant Health Inspection Service, US Department of Agriculture. HPAI confirmed cases in livestock [cited 2025 Feb 13]. https://www.aphis.usda.gov/livestock-poultry-disease/avian/avian-influenza/hpai-detections/hpai-confirmed-cases-livestock
- Ramey AM, Reeves AB, Lagassé BJ, Patil V, Hubbard LE, Kolpin DW, et al. Evidence for interannual persistence of infectious influenza A viruses in Alaska wetlands. Sci Total Environ. 2022;803:
150078 . DOIPubMedGoogle Scholar - British Columbia Ministry of Agriculture. HPAI detections in BC: wildlife and environmental surveillance. [cited 2024 Dec 2]. https://governmentofbc.maps.arcgis.com/apps/dashboards/8c6c84718e5748179102a0be2368029a
- Aoki FY, Boivin G. Influenza virus shedding: excretion patterns and effects of antiviral treatment. J Clin Virol. 2009;44:255–61. DOIPubMedGoogle Scholar