Genesis and Spread of Novel Highly Pathogenic Avian Influenza A(H5N1) Clade 2.3.4.4b Virus Genotype EA-2023-DG Reassortant, Western Europe
Steven Van Borm

, Ann Kathrin Ahrens, Claudia Bachofen, Ashley C. Banyard, Cathrine Arnason Bøe, François-Xavier Briand, Zuzana Dirbakova, Marc Engelsma, Alice Fusaro, Evelien Germeraad, Britt Gjerset, Béatrice Grasland, Frank Harders, Pierre Hostyn, Ari Kauppinen, Bénédicte Lambrecht, Benjamin C. Mollett, Isabella Monne, Alexander Nagy, Anne Pohlmann, Daniel Polzer, Scott M. Reid, Sandra Revilla-Fernandez, Mieke Steensels, Michaela Stätter, Edyta Swieton, Niina Tammiranta, Michele Wyler, Bianca Zecchin, Siamak Zohari, and Simon Dellicour
Author affiliation: Avian Virology and Immunology Unit, Sciensano, Brussels, Belgium (S. Van Borm, P. Hostyn, B. Lambrecht, M. Steensels); Institute of Diagnostic Virology, Friedrich Loeffler Institut, Greifswald-Insel Riems, Germany (A.K. Ahrens, A. Pohlmann); Institute of Virology and Immunology, Federal Department of Home Affairs, Mittelhäusern, Switzerland (C. Bachofen, M. Wyler); World Organisation for Animal Health/Food and Agriculture Organization of the United Nations International Reference Laboratory for Avian Influenza and Newcastle Disease, Animal and Plant Health Agency-Weybridge, Addlestone, UK (A.C. Banyard, B.C. Mollett, S.M. Reid); Norwegian Veterinary Institute, Ås, Norway (C. Arnason Bøe, B. Gjerset); Agence Nationale de Sécurité Sanitaire de l’alimentation, de l’environnement et du travail, Ploufragan-Plouzané-Niort, France (F.-X. Briand, B. Grasland); State Veterinary Institute Department of Animal Health, Zvolen, Slovakia (D. Dirbakova); Wageningen Bioveterinary Research Department of Virology, Lelystad, the Netherlands (M. Engelsma, E. Germeraad, F. Harders); European Reference Laboratory for Avian Influenza and Newcastle Disease, Istituto Zooprofilattico Sperimentale delle Venezie, Padua, Italy (A. Fusaro, I. Monne, B. Zecchin); Finnish Food Authority, Helsinki, Finland (A. Kauppinen, N. Tammiranta); Department of Molecular Biology, State Veterinary Institute Prague, Praha, Czech Republic (A. Nagy); Austrian Agency for Health and Food Safety, Institute for Veterinary Disease Control, Mödling, Austria (D. Polzer, S. Revilla-Fernandez, M. Stätter); National Veterinary Research Institute Department of Research Support, Puławy, Poland (E. Swieton); Swedish Veterinary Agency Department of Microbiology, Uppsala, Sweden (S. Zohari); Spatial Epidemiology Lab (SpELL), Université Libre de Bruxelles, Brussels, Belgium (S. Dellicour); Laboratory for Clinical and Epidemiological Virology, Department of Microbiology, Immunology and Transplantation, Rega Institute, KU Leuven, Leuven, Belgium (S. Dellicour); Interuniversity Institute of Bioinformatics in Brussels, Université Libre de Bruxelles, Vrije Universiteit Brussel, Brussels (S. Dellicour)
Main Article
Figure 1
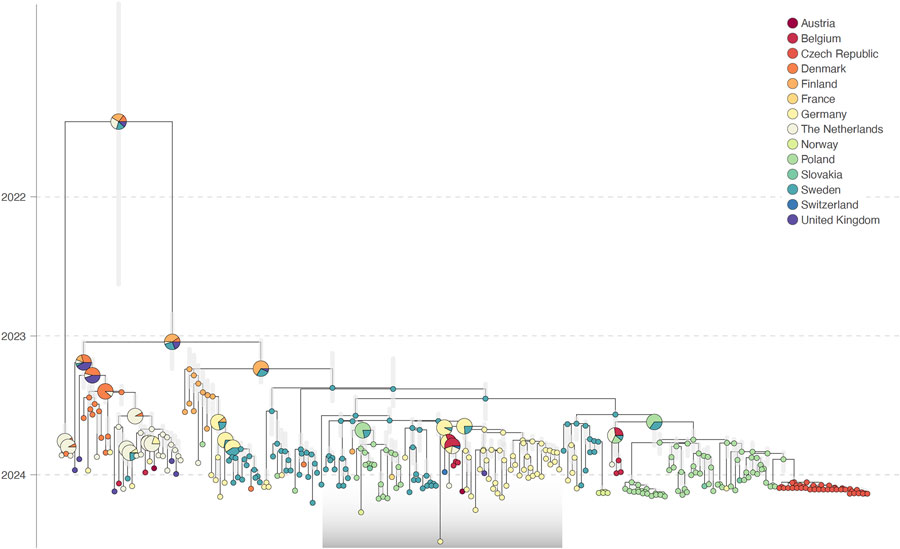
Figure 1. Discrete phylogeographic analysis from study of genesis and spread of novel highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus genotype EA-2023-DG reassortant, western Europe. The maximum clade credibility (MCC) tree was obtained from discrete phylogeographic inference based on the analysis of the PB1-HA-NP-NA-MP-NS (polymerase basic 1, hemagglutinin, nucleoprotein, neuraminidase, matrix protein, nonstructural protein) concatenated alignment of EA-2023-DG samples and selected EA-2021-AB reference sequences sourced from GISAID EpiFlu (http://www.gisaid.org). Vertical gray shaded bars reflect the 95% highest posterior density interval associated with each internal node age estimate; internal nodes are colored according to their inferred location, and tip nodes are colored according to their sampling location. For the internal nodes, when a single location could not be inferred with a posterior probability >0.95, we used a pie chart to display the posterior probabilities associated with inferred locations with a posterior probability of >0.05. The gray transparent box highlights the position of the EA-2023-DG clade. The discrete phylogeographic reconstruction based on the analysis of the polymerase basic 2 and polymerase acidic segments are available as supplementary information (Appendix 1 Figures 1, 2).
Main Article
Page created: April 21, 2025
Page updated: May 27, 2025
Page reviewed: May 27, 2025
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
