Volume 31, Number 7—July 2025
Synopsis
Human Streptococcus suis Infections, South America, 1995–2024
Figure 1
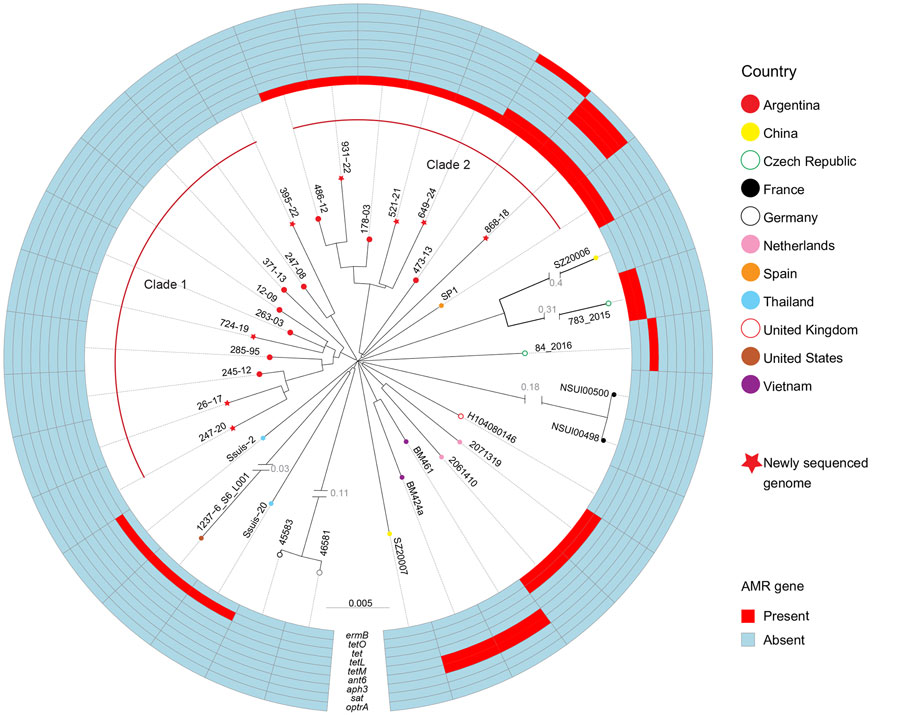
Figure 1. Phylogenetic relationships and AMR gene profiles of isolates in a study of human Streptococcus suis infections, South America, 1995–2024. Maximum-likelihood phylogenetic tree shows relationships among sequence type (ST) 1 serotype 2 S. suis isolates from human disease in Argentina and selected ST1 serotype 2 isolates from other countries. Tree was constructed by using 5,504 bp nonredundant core-genome single-nucleotide polymorphism loci identified relative to the genome sequence of ST1 serotype 2 reference strain P1/7 (not included in the depiction). The tree delineates 2 distinct clades from Argentina that differ on AMR gene content. One isolate from a human infection in Spain clusters tightly within clade 2 from Argentina, but the rest of the isolates, all from outside South America, are more distantly related genetically to either clade from Argentina. AMR genes identified in the genomes of the isolates are represented in the outer rings. AMR, antimicrobial resistance.