Volume 31, Number 7—July 2025
Dispatch
Emergence and Prevalence of Vibrio cholerae O1 Sequence Type 75 Clonal Complex, Fujian Province, China, 2009–2023
Figure
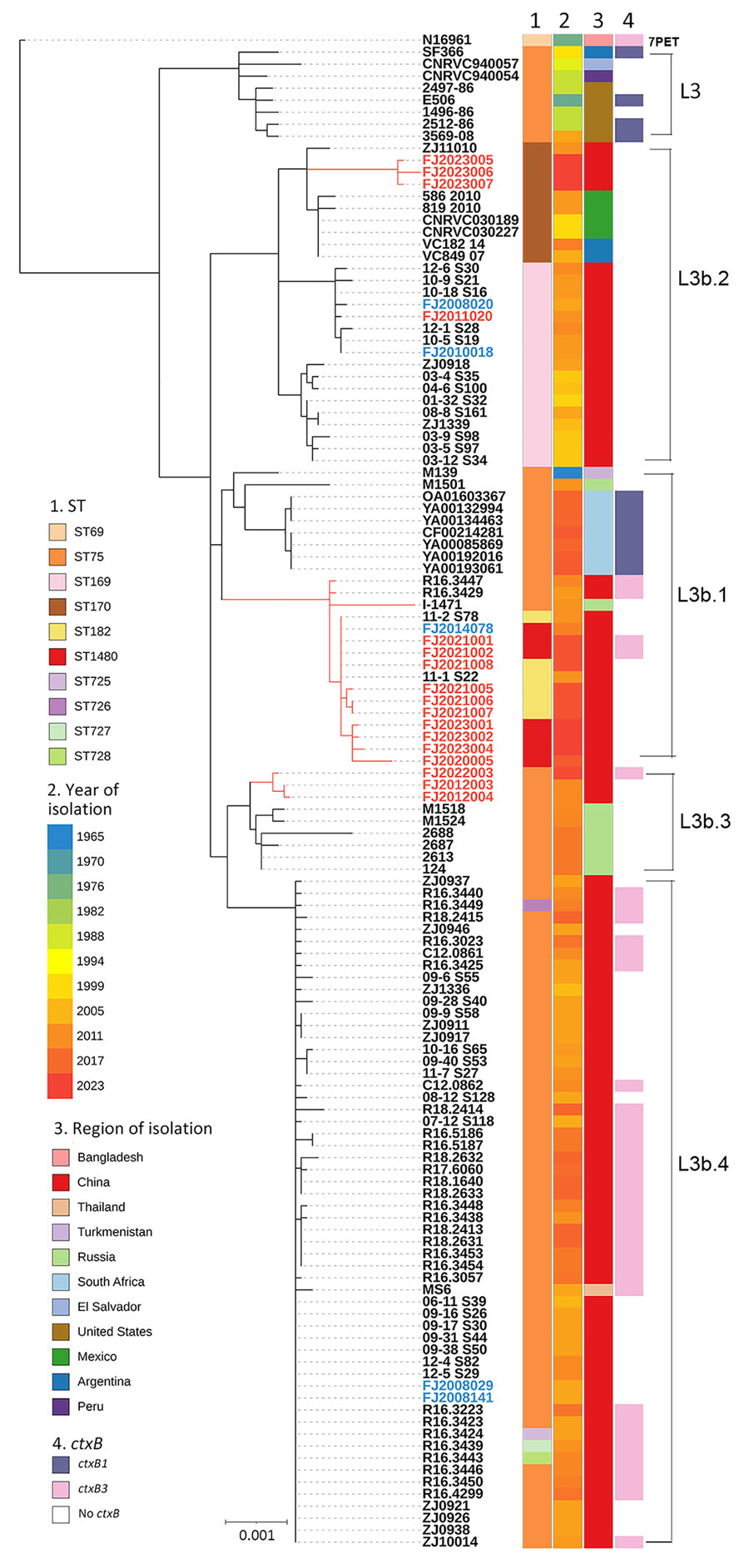
Figure. Maximum-likelihood phylogeny of genomic sequences from a study of emergence and prevalence of Vibrio cholerae O1 ST75 clonal complex, Fujian Province, China, 2009–2023. The tree revealed 5 distinct lineages: L3 (Gulf Coast), L3b.1, L3b.2, L3b.3, and L3b.4. ST1480 and ST182 clustered in L3b.1 and had a maximum pairwise distance of 0–6 SNPs. The 7PET genome N16961 (ST69) was included as an outgroup. Isolates from Fujian Province are show as red branch lines and labels for clinical isolates (n = 17) and blue text for environmental isolates (n = 5); and the other 102 isolates (black text) are of global origin. Sequence type, year and region of isolation, and types of ctxB are shown at the tips of the tree; lineages are marked on the right side of the tree. The scale bar indicates substitutions per variable site. 7PET, 7th pandemic V. cholerae O1 El Tor; ctxB, cholera toxin B subunit gene; ST, sequence type.