Volume 31, Number 7—July 2025
Research
Emergence of Distinct Salmonella enterica Serovar Enteritidis Lineage since 2020, South Korea
Figure 2
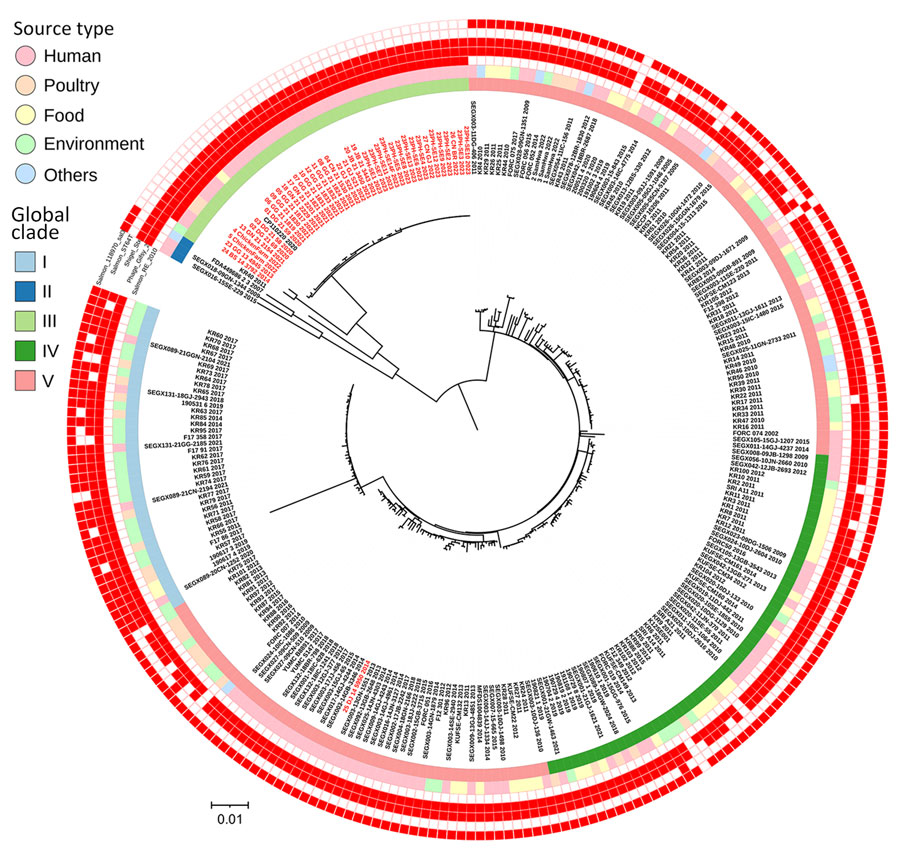
Figure 2. Phylogenetic analysis of distinct Salmonella enterica serovar Enteritidis lineage since 2020, South Korea. Maximum-likelihood phylogenetic tree was constructed for whole-genome single-nucleotide polymorphisms of Salmonella Enteritidis from South Korea (n = 223) and isolates sequenced in this study (n = 38). Red text indicates isolates from this study. Innermost ring indicates clades separated by using FastBAPS (20), followed by sources of isolates. Five outer rings indicate the presence of different prophages; red boxes indicate the presence of prophages with a total count of >30. Isolation year is indicated at the end of each isolate number. Scale bar indicates number of single-nucleotide polymorphisms per site.
References
- Ferrari RG, Rosario DKA, Cunha-Neto A, Mano SB, Figueiredo EES, Conte-Junior CA. Worldwide epidemiology of Salmonella serovars in animal-based foods: a meta-analysis. Appl Environ Microbiol. 2019;85:e00591–19. DOIPubMedGoogle Scholar
- Jeong HJ, Shin E, Park J, Han J, Kim J, Yoo J. Trends in serotype distribution of Salmonella enterica isolated from diarrheal patients in Korea, 2019 to 2021. Public Health Wkly Rep. 2022;15:2615–31.
- Hwang MJ, Park SY, Kim H, Yang SJ, Yang S, Yang JS. Waterborne and foodborne disease outbreaks in the Republic of Korea, 2023. Public Health Wkly Rep. 2025;18:17–32.
- Eun JS, Han J, Lim JH, Shin E, Kim J, Ko DJ, et al. Salmonellosis outbreaks linked to eggs at 2 gimbap restaurants in Korea. Epidemiol Health. 2024;46:
e2024036 . DOIPubMedGoogle Scholar - European Centre for Disease Prevention and Control, European Food Safety Authority. Multi‐country outbreak of Salmonella Enteritidis sequence type (ST)11 infections linked to eggs and egg products—8 February 2022. EFSA Support Publ. 2022;19:7180E.
- Benson HE, Reeve L, Findlater L, Vusirikala A, Pietzsch M, Olufon O, et al.; Incident Management Team. Local Salmonella Enteritidis restaurant outbreak investigation in England provides further evidence for eggs as source in widespread international cluster, March to April 2023. Euro Surveill. 2023;28:
2300309 . DOIPubMedGoogle Scholar - Zhou Z, Alikhan NF, Mohamed K, Fan Y, Achtman M; Agama Study Group. The EnteroBase user’s guide, with case studies on Salmonella transmissions, Yersinia pestis phylogeny, and Escherichia core genomic diversity. Genome Res. 2020;30:138–52. DOIPubMedGoogle Scholar
- Zhou Z, Charlesworth J, Achtman M. HierCC: a multi-level clustering scheme for population assignments based on core genome MLST. Bioinformatics. 2021;37:3645–6. DOIPubMedGoogle Scholar
- La TM, Kim T, Lee HJ, Lee JB, Park SY, Choi IS, et al. Whole-genome analysis of multidrug-resistant Salmonella Enteritidis strains isolated from poultry sources in Korea. Pathogens. 2021;10:1615. DOIPubMedGoogle Scholar
- Hyeon JY, Li S, Mann DA, Zhang S, Kim KJ, Lee DH, et al. Whole-genome sequencing analysis of Salmonella enterica serotype Enteritidis isolated from poultry sources in South Korea, 2010–2017. Pathogens. 2021;10:45. DOIPubMedGoogle Scholar
- Jang HJ, Ha YK, Yu SN, Kim SY, Um J, Ha GJ, et al. Molecular epidemiological analysis of food poisoning caused by Salmonella enterica serotype Enteritidis in Gyeongnam Province of Korea. J Life Sci. 2023;33:56–63.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. DOIPubMedGoogle Scholar
- Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–4. DOIPubMedGoogle Scholar
- Liu B, Zheng D, Zhou S, Chen L, Yang J. VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 2022;50(D1):D912–7. DOIPubMedGoogle Scholar
- Wishart DS, Han S, Saha S, Oler E, Peters H, Grant JR, et al. PHASTEST: faster than PHASTER, better than PHAST. Nucleic Acids Res. 2023;51(W1):W443–50. DOIPubMedGoogle Scholar
- Li S, He Y, Mann DA, Deng X. Global spread of Salmonella Enteritidis via centralized sourcing and international trade of poultry breeding stocks. Nat Commun. 2021;12:5109. DOIPubMedGoogle Scholar
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: A fast and versatile genome alignment system. PLOS Comput Biol. 2018;14:
e1005944 . DOIPubMedGoogle Scholar - Croucher NJ, Page AJ, Connor TR, Delaney AJ, Keane JA, Bentley SD, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:
e15 . DOIPubMedGoogle Scholar - Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–74. DOIPubMedGoogle Scholar
- Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Res. 2019;47:5539–49. DOIPubMedGoogle Scholar
- Katz LS, Griswold T, Morrison SS, Caravas JA, Zhang S, den Bakker HC, et al. Mashtree: a rapid comparison of whole genome sequence files. J Open Source Softw. 2019;4:1762. DOIPubMedGoogle Scholar
- Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016;2:
vew007 . DOIPubMedGoogle Scholar - Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4:
vey016 . DOIPubMedGoogle Scholar - Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 2020;37:291–4. DOIPubMedGoogle Scholar
- Xie W, Lewis PO, Fan Y, Kuo L, Chen MH. Improving marginal likelihood estimation for Bayesian phylogenetic model selection. Syst Biol. 2011;60:150–60. DOIPubMedGoogle Scholar
- Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol. 2016;33:2167–9. DOIPubMedGoogle Scholar
- Hajra D, Nair AV, Chakravortty D. Decoding the invasive nature of a tropical pathogen of concern: The invasive non-Typhoidal Salmonella strains causing host-restricted extraintestinal infections worldwide. Microbiol Res. 2023;277:
127488 . DOIPubMedGoogle Scholar - Food and Agriculture Organization of the United Nations. Food and agriculture data, 2025 [cited 2025 May 5]. https://www.fao.org/faostat
- European Centre for Disease Prevention and Control. European Food Safety Authority. Multi‐country outbreak of Salmonella Enteritidis sequence type (ST)11 infections linked to poultry products in the EU/EEA and the United Kingdom. EFSA Support Publ. 2021;18:6486E.
1These first authors contributed equally to this article.
2These senior authors contributed equally to this article.