Volume 31, Number 7—July 2025
Synopsis
Community Outbreak of OXA-48–Producing Escherichia coli Linked to Food Premises, New Zealand, 2018–2022
Figure 2
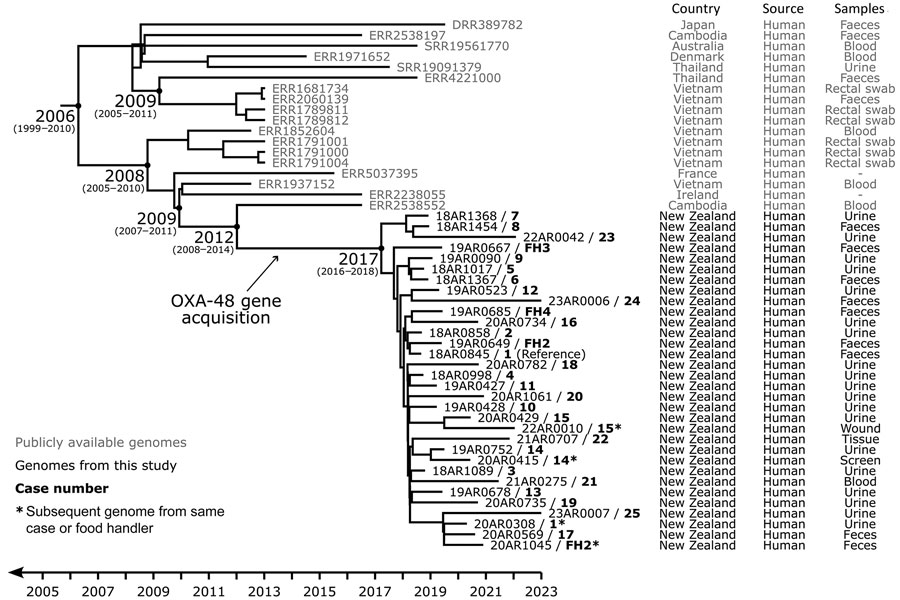
Figure 2. Evolutionary reconstruction for OXA-48–producing Escherichia coli sequence type (ST)131 genomes obtained from cases and food handlers compared with publicly available genomes in study of community outbreak linked to food premises, Hutt Valley, New Zealand, August 2018–December 2022. A time-calibrated maximum clade credibility tree was inferred from 323 nonrecombinant orthologous biallelic core-genome single-nucleotide variants (SNVs) from 50 ST131 genomes. SNVs were derived from a core-genome alignment of ≈4,767,900 bp and were called against the chromosome of 18AR0845 (GenBank accession no. CP175691). The x-axis represents the emergence time estimates. Case numbers (1–25), shown in bold after the genome codes, correspond to case reference numbers shown in Appendix Table 2). Case numbers FH1–4 indicate genomes obtained from food handlers working at a community-based food premises to which 18 of the case-patients had been exposed. Asterisks indicate subsequent genomes obtained from the same case-patient or food handler.