Volume 31, Number 8—August 2025
CME ACTIVITY - Synopsis
Emergence of Clade Ib Monkeypox Virus—Current State of Evidence
Figure 2
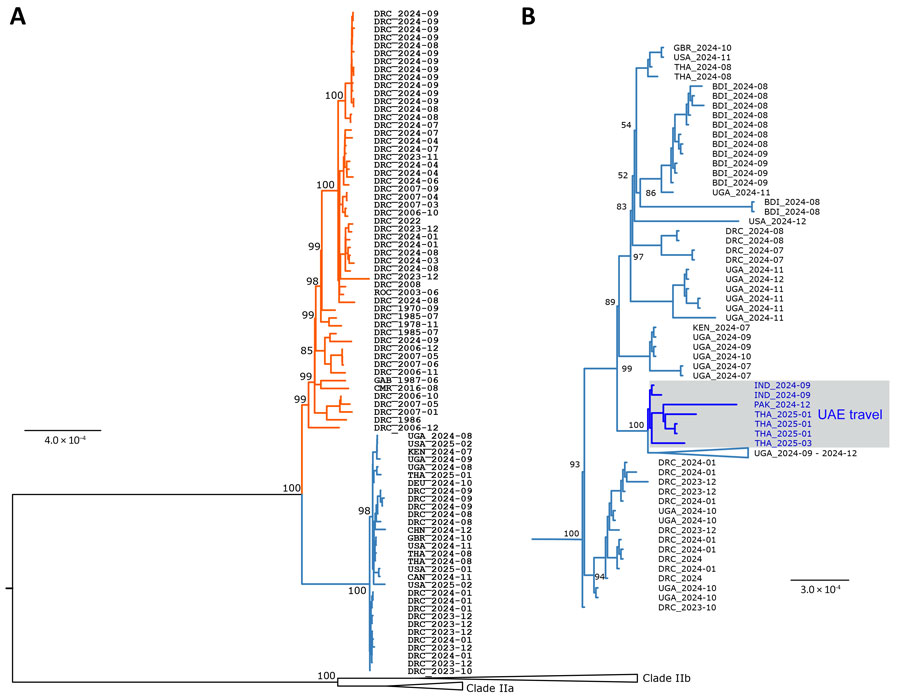
Figure 2. Phylogenetic analysis of clade I monkeypox virus (MPXV) sequences. A) Phylogenetic tree estimated for sequences from 1970–2024 according to core single-nucleotide differences among 103 clade I MPXV genomes. Sequences are identified by country (3-letter code) and collection date (year-month). Red branches indicate MPXV clade Ia; blue branches indicate clade Ib. Numbers at branch points indicate bootstrap support values (percentage of 1,000 replicates). B) Phylogenetic analysis of select clade Ib MPXV sequences. Blue text in gray box indicates sequences linked to persons who had traveled to UAE. Phylogenetic analyses were performed by using the maximum-likelihood method and the 3 substitution types model and equal base frequency plus empirical base frequencies plus proportion of invariable sites substitution model in ModelFinder (IQ-TREE version 2.2.6, https://iqtree.github.io). Whole-genome alignments of sequences downloaded from GISAID (https://www.gisaid.org) and GenBank (Appendix) databases were generated by using MAFFT version 7.490 (https://mafft.cbrc.jp/alignment/software). Trees were rooted by using clade II reference genomes: ON627808, ON585033, DQ011156, and DQ011153. Clade Ia sequences are not shown in the tree in panel B. Scale bars indicate nucleotide substitutions per site. BDI, Burundi; CAN, Canada; CHN, China; CMR, Cameroon; DEU, Germany; DRC, Democratic Republic of the Congo; GAB, Gabon; GBR, United Kingdom; IND, India; KEN, Kenya; PAK, Pakistan; ROC, Republic of Congo; THA, Thailand; UAE, United Arab Emirates; UGA, Uganda; USA, United States.
1These first authors contributed equally to the article.